Aeromonas hydrophila ATCC 7966
Lineage
› cellular organisms
› Bacteria
› Proteobacteria
› Gammaproteobacteria
› Aeromonadales
› Aeromonadaceae
› Aeromonas
› Aeromonas hydrophila
› Aeromonas hydrophila subsp. hydrophila
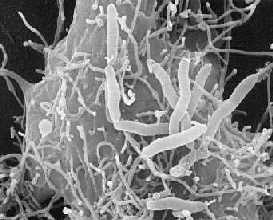
Aeromonas hydrophila
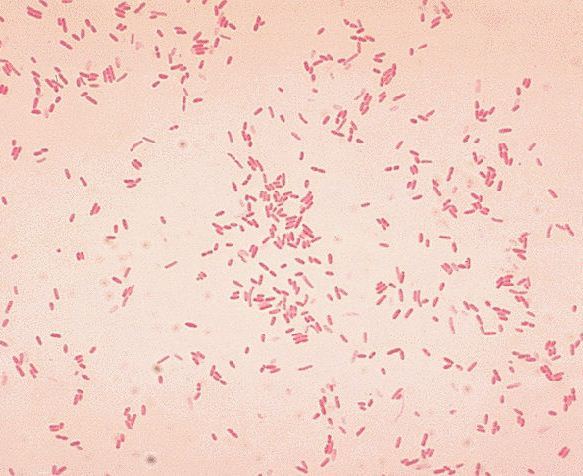
Aeromonas hydrophila
Description
Aeromonas hydrophila is a heterotrophic, Gram-negative, rod shaped bacterium, mainly found in areas with a warm climate. This bacterium can also be found in fresh, salt, marine, estuarine, chlorinated, and un-chlorinated water. A. hydrophila can survive in aerobic and anaerobic environments. This bacterium can digest materials such as gelatin, and hemoglobin. A. hydrophila was isolated from humans and animals in the 1950s. This bacterium is the most well known of the six species of Aeromonas. It is also highly resistant to multiple medications, chlorine, and cold temperatures.
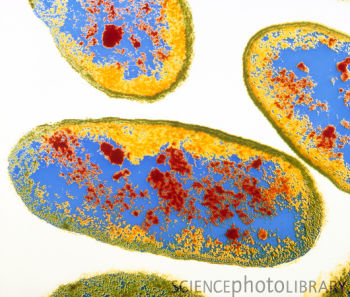
Aeromonas hydrophila
A. hydrophila is a facultative anaerobic invasive motile gram-negative bacillus that commonly found in water. This pathogenic bacterium is reported to cause water-associated traumatic secondary wound infections, ulcerative disease syndrome in fish, septicemia, cellulitis, pneumonitis, necrotizing fasciitis, and gasteroenteritis. À. hydrophila is associated with 100% mortality for septic shock.
Antibiotic therapy is generally recommended and Cefotaxime and other third-generation cephalosporins may be an effective treatment option. No vaccine is available so far.
Sequencing Aeromonas hydrophila subsp. hydrophila ATCC 7966 was carried out and completed on November 8, 2006 by TIGR. The circular genome size is 4744448 bp consisting of one circular chromosome that encodes 4122 proteins.
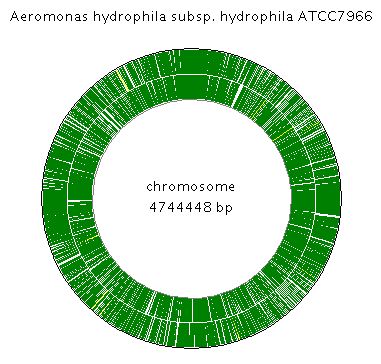
Circular chromosome
PubMed database
In the PubMed database you can find
(searching for Aeromonas hydrophila ATCC[TI], for example)
an interesting article Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades.
(PMID: 16980456; by Seshadri R, Joseph SW, Chopra AK and others.)
Here is its abstract::
The complete genome of Aeromonas hydrophila ÀÒÑÑ 7966 (T) was sequenced. Aeromonas, a ubiquitous waterborne bacterium, has been placed by the Environmental Protection Agency on the Contaminant Candidate List because of its potential to cause human disease. The 4.7-Mb genome of this emerging pathogen shows a physiologically adroit organism with broad metabolic capabilities and considerable virulence potential. A large array of virulence genes, including some identified in clinical isolates of Aeromonas spp. or Vibrio spp., may confer upon this organism the ability to infect a wide range of hosts. However, two recognized virulence markers, a type III secretion system and a lateral flagellum, that are reported in other A. hydrophila strains are not identified in the sequenced isolate, ATCC 7966(T). Given the ubiquity and free-living lifestyle of this organism, there is relatively little evidence of fluidity in terms of mobile elements in the genome of this particular strain. Notable aspects of the metabolic repertoire of À. hydrophila include dissimilatory sulfate reduction and resistance mechanisms (such as thiopurine reductase, arsenate reductase, and phosphonate degradation enzymes) against toxic compounds encountered in polluted waters. These enzymes may have bioremediative as well as industrial potential. Thus, Thus, the À. hydrophila genome sequence provides valuable insights into its ability to flourish in both aquatic and host environments.
You can read the full article in the Journal of Bacteriology.
And the identification of the special secretion system of À. hydrophila may facilitate to develop suitable vaccines as well as to understand more of the pathogenesis of A. hydrophila.
Extra information
| Protein count | 4122 |
| Length (bp) | 4744448 |
| Av. CDS Length | 1007.771 |
| GC content | 61.5% |
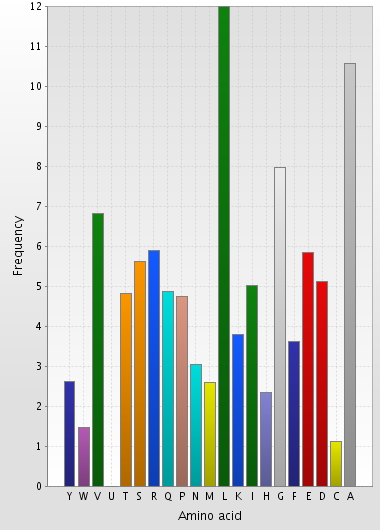
Amino acid composition
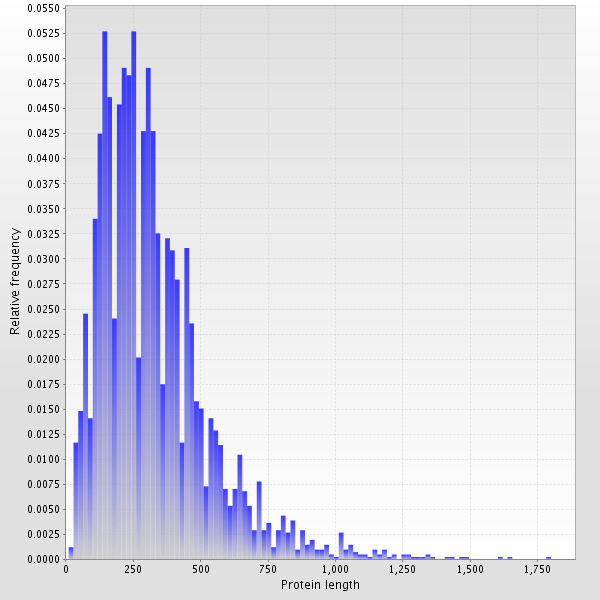
Protein length distribution
You can find the complete genome on the webpages of European Bioinformatics Institute and The National Center for Biotechnology Information.
Links
Sources
- Bioinformatics Resource Portal
- Genomic Target Database
- Wikipedia
- Microbe Wiki
- The Universal Protein Resource
- PubMed
- European Bioinformatics Institute
Sources of images
See also
- University of Lausanne
- The Microbial Signal Transduction database
- Bioinformatic tools server
- DNA Data Bank of Japan
- Carbohydrate-Active enZYmes Database
- StrainInfo bioportal
- The National Center for Biotechnology Information
- European Bioinformatics Institute
- SUPERFAMILY
- Journal of Bacteriology
- Pathosystems Resource Integration Center
- Prokaryotic 2-Component Systems
- European Bioinformatics Institute
- Microbial Genome Browser
