:-) G R O M A C S (-:
S C A M O R G
:-) VERSION 4.5.5 (-:
Written by Emile Apol, Rossen Apostolov, Herman J.C. Berendsen,
Aldert van Buuren, Pär Bjelkmar, Rudi van Drunen, Anton Feenstra,
Gerrit Groenhof, Peter Kasson, Per Larsson, Pieter Meulenhoff,
Teemu Murtola, Szilard Pall, Sander Pronk, Roland Schulz,
Michael Shirts, Alfons Sijbers, Peter Tieleman,
Berk Hess, David van der Spoel, and Erik Lindahl.
Copyright (c) 1991-2000, University of Groningen, The Netherlands.
Copyright (c) 2001-2010, The GROMACS development team at
Uppsala University & The Royal Institute of Technology, Sweden.
check out http://www.gromacs.org for more information.
This program is free software; you can redistribute it and/or
modify it under the terms of the GNU General Public License
as published by the Free Software Foundation; either version 2
of the License, or (at your option) any later version.
:-) mdrun (-:
Option Filename Type Description
------------------------------------------------------------
-s et_be.tpr Input Run input file: tpr tpb tpa
-o et_be.tpr.trr Output Full precision trajectory: trr trj cpt
-x et_be.tpr.xtc Output, Opt. Compressed trajectory (portable xdr format)
-cpi et_be.tpr.cpt Input, Opt. Checkpoint file
-cpo et_be.tpr.cpt Output, Opt. Checkpoint file
-c et_be.tpr.gro Output Structure file: gro g96 pdb etc.
-e et_be.tpr.edr Output Energy file
-g et_be.tpr.log Output Log file
-dhdl et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-field et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-table et_be.tpr.xvg Input, Opt. xvgr/xmgr file
-tablep et_be.tpr.xvg Input, Opt. xvgr/xmgr file
-tableb et_be.tpr.xvg Input, Opt. xvgr/xmgr file
-rerun et_be.tpr.trr Input, Opt. Trajectory: xtc trr trj gro g96 pdb cpt
-tpi et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-tpid et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-ei et_be.tpr.edi Input, Opt. ED sampling input
-eo et_be.tpr.edo Output, Opt. ED sampling output
-j et_be.tpr.gct Input, Opt. General coupling stuff
-jo et_be.tpr.gct Output, Opt. General coupling stuff
-ffout et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-devout et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-runav et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-px et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-pf et_be.tpr.xvg Output, Opt. xvgr/xmgr file
-mtx et_be.tpr.mtx Output, Opt. Hessian matrix
-dn et_be.tpr.ndx Output, Opt. Index file
-multidir et_be.tpr Input, Opt., Mult. Run directory
Option Type Value Description
------------------------------------------------------
-[no]h bool no Print help info and quit
-[no]version bool no Print version info and quit
-nice int 0 Set the nicelevel
-deffnm string et_be.tpr Set the default filename for all file options
-xvg enum xmgrace xvg plot formatting: xmgrace, xmgr or none
-[no]pd bool no Use particle decompostion
-dd vector 0 0 0 Domain decomposition grid, 0 is optimize
-nt int 1 Number of threads to start (0 is guess)
-npme int -1 Number of separate nodes to be used for PME, -1
is guess
-ddorder enum interleave DD node order: interleave, pp_pme or cartesian
-[no]ddcheck bool yes Check for all bonded interactions with DD
-rdd real 0 The maximum distance for bonded interactions with
DD (nm), 0 is determine from initial coordinates
-rcon real 0 Maximum distance for P-LINCS (nm), 0 is estimate
-dlb enum auto Dynamic load balancing (with DD): auto, no or yes
-dds real 0.8 Minimum allowed dlb scaling of the DD cell size
-gcom int -1 Global communication frequency
-[no]v bool yes Be loud and noisy
-[no]compact bool yes Write a compact log file
-[no]seppot bool no Write separate V and dVdl terms for each
interaction type and node to the log file(s)
-pforce real -1 Print all forces larger than this (kJ/mol nm)
-[no]reprod bool no Try to avoid optimizations that affect binary
reproducibility
-cpt real 15 Checkpoint interval (minutes)
-[no]cpnum bool no Keep and number checkpoint files
-[no]append bool yes Append to previous output files when continuing
from checkpoint instead of adding the simulation
part number to all file names
-maxh real -1 Terminate after 0.99 times this time (hours)
-multi int 0 Do multiple simulations in parallel
-replex int 0 Attempt replica exchange periodically with this
period (steps)
-reseed int -1 Seed for replica exchange, -1 is generate a seed
-[no]ionize bool no Do a simulation including the effect of an X-Ray
bombardment on your system
Back Off! I just backed up et_be.tpr.log to ./#et_be.tpr.log.2#
Getting Loaded...
Reading file et_be.tpr, VERSION 4.5.5 (single precision)
Loaded with Money
Back Off! I just backed up et_be.tpr.trr to ./#et_be.tpr.trr.2#
Back Off! I just backed up et_be.tpr.edr to ./#et_be.tpr.edr.2#
starting mdrun 'first one'
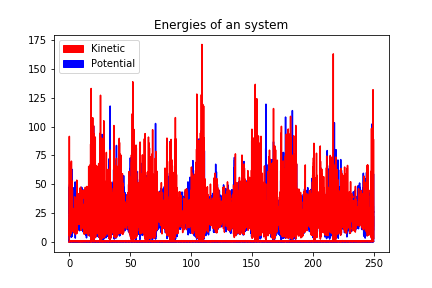
250000 steps, 250.0 ps.
step 249900, remaining runtime: 0 s
Writing final coordinates.
Back Off! I just backed up et_be.tpr.gro to ./#et_be.tpr.gro.2#
step 250000, remaining runtime: 0 s
NODE (s) Real (s) (%)
Time: 3.890 4.147 93.8
(Mnbf/s) (MFlops) (ns/day) (hour/ns)
Performance: 0.000 350.890 5552.722 0.004
gcq#15: "Don't Grumble, Give a Whistle !" (Monty Python)
:-) G R O M A C S (-:
GRoups of Organic Molecules in ACtion for Science
:-) VERSION 4.5.5 (-:
Written by Emile Apol, Rossen Apostolov, Herman J.C. Berendsen,
Aldert van Buuren, Pär Bjelkmar, Rudi van Drunen, Anton Feenstra,
Gerrit Groenhof, Peter Kasson, Per Larsson, Pieter Meulenhoff,
Teemu Murtola, Szilard Pall, Sander Pronk, Roland Schulz,
Michael Shirts, Alfons Sijbers, Peter Tieleman,
Berk Hess, David van der Spoel, and Erik Lindahl.
Copyright (c) 1991-2000, University of Groningen, The Netherlands.
Copyright (c) 2001-2010, The GROMACS development team at
Uppsala University & The Royal Institute of Technology, Sweden.
check out http://www.gromacs.org for more information.
This program is free software; you can redistribute it and/or
modify it under the terms of the GNU General Public License
as published by the Free Software Foundation; either version 2
of the License, or (at your option) any later version.
:-) mdrun (-:
Option Filename Type Description
------------------------------------------------------------
-s et_vr.tpr Input Run input file: tpr tpb tpa
-o et_vr.tpr.trr Output Full precision trajectory: trr trj cpt
-x et_vr.tpr.xtc Output, Opt. Compressed trajectory (portable xdr format)
-cpi et_vr.tpr.cpt Input, Opt. Checkpoint file
-cpo et_vr.tpr.cpt Output, Opt. Checkpoint file
-c et_vr.tpr.gro Output Structure file: gro g96 pdb etc.
-e et_vr.tpr.edr Output Energy file
-g et_vr.tpr.log Output Log file
-dhdl et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-field et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-table et_vr.tpr.xvg Input, Opt. xvgr/xmgr file
-tablep et_vr.tpr.xvg Input, Opt. xvgr/xmgr file
-tableb et_vr.tpr.xvg Input, Opt. xvgr/xmgr file
-rerun et_vr.tpr.trr Input, Opt. Trajectory: xtc trr trj gro g96 pdb cpt
-tpi et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-tpid et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-ei et_vr.tpr.edi Input, Opt. ED sampling input
-eo et_vr.tpr.edo Output, Opt. ED sampling output
-j et_vr.tpr.gct Input, Opt. General coupling stuff
-jo et_vr.tpr.gct Output, Opt. General coupling stuff
-ffout et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-devout et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-runav et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-px et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-pf et_vr.tpr.xvg Output, Opt. xvgr/xmgr file
-mtx et_vr.tpr.mtx Output, Opt. Hessian matrix
-dn et_vr.tpr.ndx Output, Opt. Index file
-multidir et_vr.tpr Input, Opt., Mult. Run directory
Option Type Value Description
------------------------------------------------------
-[no]h bool no Print help info and quit
-[no]version bool no Print version info and quit
-nice int 0 Set the nicelevel
-deffnm string et_vr.tpr Set the default filename for all file options
-xvg enum xmgrace xvg plot formatting: xmgrace, xmgr or none
-[no]pd bool no Use particle decompostion
-dd vector 0 0 0 Domain decomposition grid, 0 is optimize
-nt int 1 Number of threads to start (0 is guess)
-npme int -1 Number of separate nodes to be used for PME, -1
is guess
-ddorder enum interleave DD node order: interleave, pp_pme or cartesian
-[no]ddcheck bool yes Check for all bonded interactions with DD
-rdd real 0 The maximum distance for bonded interactions with
DD (nm), 0 is determine from initial coordinates
-rcon real 0 Maximum distance for P-LINCS (nm), 0 is estimate
-dlb enum auto Dynamic load balancing (with DD): auto, no or yes
-dds real 0.8 Minimum allowed dlb scaling of the DD cell size
-gcom int -1 Global communication frequency
-[no]v bool yes Be loud and noisy
-[no]compact bool yes Write a compact log file
-[no]seppot bool no Write separate V and dVdl terms for each
interaction type and node to the log file(s)
-pforce real -1 Print all forces larger than this (kJ/mol nm)
-[no]reprod bool no Try to avoid optimizations that affect binary
reproducibility
-cpt real 15 Checkpoint interval (minutes)
-[no]cpnum bool no Keep and number checkpoint files
-[no]append bool yes Append to previous output files when continuing
from checkpoint instead of adding the simulation
part number to all file names
-maxh real -1 Terminate after 0.99 times this time (hours)
-multi int 0 Do multiple simulations in parallel
-replex int 0 Attempt replica exchange periodically with this
period (steps)
-reseed int -1 Seed for replica exchange, -1 is generate a seed
-[no]ionize bool no Do a simulation including the effect of an X-Ray
bombardment on your system
Back Off! I just backed up et_vr.tpr.log to ./#et_vr.tpr.log.2#
Getting Loaded...
Reading file et_vr.tpr, VERSION 4.5.5 (single precision)
Loaded with Money
Back Off! I just backed up et_vr.tpr.trr to ./#et_vr.tpr.trr.2#
Back Off! I just backed up et_vr.tpr.edr to ./#et_vr.tpr.edr.2#
starting mdrun 'first one'
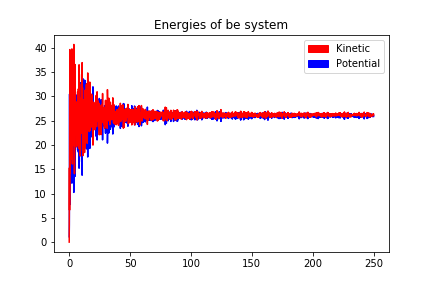
250000 steps, 250.0 ps.
step 249900, remaining runtime: 0 s
Writing final coordinates.
Back Off! I just backed up et_vr.tpr.gro to ./#et_vr.tpr.gro.2#
step 250000, remaining runtime: 0 s
NODE (s) Real (s) (%)
Time: 3.840 4.006 95.9
(Mnbf/s) (MFlops) (ns/day) (hour/ns)
Performance: 0.000 355.393 5625.023 0.004
gcq#109: "Ease Myself Into the Body Bag" (P.J. Harvey)
:-) G R O M A C S (-:
GROtesk MACabre and Sinister
:-) VERSION 4.5.5 (-:
Written by Emile Apol, Rossen Apostolov, Herman J.C. Berendsen,
Aldert van Buuren, Pär Bjelkmar, Rudi van Drunen, Anton Feenstra,
Gerrit Groenhof, Peter Kasson, Per Larsson, Pieter Meulenhoff,
Teemu Murtola, Szilard Pall, Sander Pronk, Roland Schulz,
Michael Shirts, Alfons Sijbers, Peter Tieleman,
Berk Hess, David van der Spoel, and Erik Lindahl.
Copyright (c) 1991-2000, University of Groningen, The Netherlands.
Copyright (c) 2001-2010, The GROMACS development team at
Uppsala University & The Royal Institute of Technology, Sweden.
check out http://www.gromacs.org for more information.
This program is free software; you can redistribute it and/or
modify it under the terms of the GNU General Public License
as published by the Free Software Foundation; either version 2
of the License, or (at your option) any later version.
:-) mdrun (-:
Option Filename Type Description
------------------------------------------------------------
-s et_nh.tpr Input Run input file: tpr tpb tpa
-o et_nh.tpr.trr Output Full precision trajectory: trr trj cpt
-x et_nh.tpr.xtc Output, Opt. Compressed trajectory (portable xdr format)
-cpi et_nh.tpr.cpt Input, Opt. Checkpoint file
-cpo et_nh.tpr.cpt Output, Opt. Checkpoint file
-c et_nh.tpr.gro Output Structure file: gro g96 pdb etc.
-e et_nh.tpr.edr Output Energy file
-g et_nh.tpr.log Output Log file
-dhdl et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-field et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-table et_nh.tpr.xvg Input, Opt. xvgr/xmgr file
-tablep et_nh.tpr.xvg Input, Opt. xvgr/xmgr file
-tableb et_nh.tpr.xvg Input, Opt. xvgr/xmgr file
-rerun et_nh.tpr.trr Input, Opt. Trajectory: xtc trr trj gro g96 pdb cpt
-tpi et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-tpid et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-ei et_nh.tpr.edi Input, Opt. ED sampling input
-eo et_nh.tpr.edo Output, Opt. ED sampling output
-j et_nh.tpr.gct Input, Opt. General coupling stuff
-jo et_nh.tpr.gct Output, Opt. General coupling stuff
-ffout et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-devout et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-runav et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-px et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-pf et_nh.tpr.xvg Output, Opt. xvgr/xmgr file
-mtx et_nh.tpr.mtx Output, Opt. Hessian matrix
-dn et_nh.tpr.ndx Output, Opt. Index file
-multidir et_nh.tpr Input, Opt., Mult. Run directory
Option Type Value Description
------------------------------------------------------
-[no]h bool no Print help info and quit
-[no]version bool no Print version info and quit
-nice int 0 Set the nicelevel
-deffnm string et_nh.tpr Set the default filename for all file options
-xvg enum xmgrace xvg plot formatting: xmgrace, xmgr or none
-[no]pd bool no Use particle decompostion
-dd vector 0 0 0 Domain decomposition grid, 0 is optimize
-nt int 1 Number of threads to start (0 is guess)
-npme int -1 Number of separate nodes to be used for PME, -1
is guess
-ddorder enum interleave DD node order: interleave, pp_pme or cartesian
-[no]ddcheck bool yes Check for all bonded interactions with DD
-rdd real 0 The maximum distance for bonded interactions with
DD (nm), 0 is determine from initial coordinates
-rcon real 0 Maximum distance for P-LINCS (nm), 0 is estimate
-dlb enum auto Dynamic load balancing (with DD): auto, no or yes
-dds real 0.8 Minimum allowed dlb scaling of the DD cell size
-gcom int -1 Global communication frequency
-[no]v bool yes Be loud and noisy
-[no]compact bool yes Write a compact log file
-[no]seppot bool no Write separate V and dVdl terms for each
interaction type and node to the log file(s)
-pforce real -1 Print all forces larger than this (kJ/mol nm)
-[no]reprod bool no Try to avoid optimizations that affect binary
reproducibility
-cpt real 15 Checkpoint interval (minutes)
-[no]cpnum bool no Keep and number checkpoint files
-[no]append bool yes Append to previous output files when continuing
from checkpoint instead of adding the simulation
part number to all file names
-maxh real -1 Terminate after 0.99 times this time (hours)
-multi int 0 Do multiple simulations in parallel
-replex int 0 Attempt replica exchange periodically with this
period (steps)
-reseed int -1 Seed for replica exchange, -1 is generate a seed
-[no]ionize bool no Do a simulation including the effect of an X-Ray
bombardment on your system
Back Off! I just backed up et_nh.tpr.log to ./#et_nh.tpr.log.2#
Getting Loaded...
Reading file et_nh.tpr, VERSION 4.5.5 (single precision)
Loaded with Money
Back Off! I just backed up et_nh.tpr.trr to ./#et_nh.tpr.trr.2#
Back Off! I just backed up et_nh.tpr.edr to ./#et_nh.tpr.edr.2#
starting mdrun 'first one'
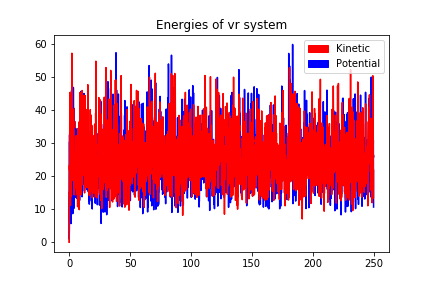
250000 steps, 250.0 ps.
step 249900, remaining runtime: 0 s
Writing final coordinates.
Back Off! I just backed up et_nh.tpr.gro to ./#et_nh.tpr.gro.2#
step 250000, remaining runtime: 0 s
NODE (s) Real (s) (%)
Time: 3.870 4.038 95.8
(Mnbf/s) (MFlops) (ns/day) (hour/ns)
Performance: 0.000 352.435 5581.418 0.004
gcq#203: "Hangout In the Suburbs If You've Got the Guts" (Urban Dance Squad)
:-) G R O M A C S (-:
Glycine aRginine prOline Methionine Alanine Cystine Serine
:-) VERSION 4.5.5 (-:
Written by Emile Apol, Rossen Apostolov, Herman J.C. Berendsen,
Aldert van Buuren, Pär Bjelkmar, Rudi van Drunen, Anton Feenstra,
Gerrit Groenhof, Peter Kasson, Per Larsson, Pieter Meulenhoff,
Teemu Murtola, Szilard Pall, Sander Pronk, Roland Schulz,
Michael Shirts, Alfons Sijbers, Peter Tieleman,
Berk Hess, David van der Spoel, and Erik Lindahl.
Copyright (c) 1991-2000, University of Groningen, The Netherlands.
Copyright (c) 2001-2010, The GROMACS development team at
Uppsala University & The Royal Institute of Technology, Sweden.
check out http://www.gromacs.org for more information.
This program is free software; you can redistribute it and/or
modify it under the terms of the GNU General Public License
as published by the Free Software Foundation; either version 2
of the License, or (at your option) any later version.
:-) mdrun (-:
Option Filename Type Description
------------------------------------------------------------
-s et_an.tpr Input Run input file: tpr tpb tpa
-o et_an.tpr.trr Output Full precision trajectory: trr trj cpt
-x et_an.tpr.xtc Output, Opt. Compressed trajectory (portable xdr format)
-cpi et_an.tpr.cpt Input, Opt. Checkpoint file
-cpo et_an.tpr.cpt Output, Opt. Checkpoint file
-c et_an.tpr.gro Output Structure file: gro g96 pdb etc.
-e et_an.tpr.edr Output Energy file
-g et_an.tpr.log Output Log file
-dhdl et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-field et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-table et_an.tpr.xvg Input, Opt. xvgr/xmgr file
-tablep et_an.tpr.xvg Input, Opt. xvgr/xmgr file
-tableb et_an.tpr.xvg Input, Opt. xvgr/xmgr file
-rerun et_an.tpr.trr Input, Opt. Trajectory: xtc trr trj gro g96 pdb cpt
-tpi et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-tpid et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-ei et_an.tpr.edi Input, Opt. ED sampling input
-eo et_an.tpr.edo Output, Opt. ED sampling output
-j et_an.tpr.gct Input, Opt. General coupling stuff
-jo et_an.tpr.gct Output, Opt. General coupling stuff
-ffout et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-devout et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-runav et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-px et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-pf et_an.tpr.xvg Output, Opt. xvgr/xmgr file
-mtx et_an.tpr.mtx Output, Opt. Hessian matrix
-dn et_an.tpr.ndx Output, Opt. Index file
-multidir et_an.tpr Input, Opt., Mult. Run directory
Option Type Value Description
------------------------------------------------------
-[no]h bool no Print help info and quit
-[no]version bool no Print version info and quit
-nice int 0 Set the nicelevel
-deffnm string et_an.tpr Set the default filename for all file options
-xvg enum xmgrace xvg plot formatting: xmgrace, xmgr or none
-[no]pd bool no Use particle decompostion
-dd vector 0 0 0 Domain decomposition grid, 0 is optimize
-nt int 1 Number of threads to start (0 is guess)
-npme int -1 Number of separate nodes to be used for PME, -1
is guess
-ddorder enum interleave DD node order: interleave, pp_pme or cartesian
-[no]ddcheck bool yes Check for all bonded interactions with DD
-rdd real 0 The maximum distance for bonded interactions with
DD (nm), 0 is determine from initial coordinates
-rcon real 0 Maximum distance for P-LINCS (nm), 0 is estimate
-dlb enum auto Dynamic load balancing (with DD): auto, no or yes
-dds real 0.8 Minimum allowed dlb scaling of the DD cell size
-gcom int -1 Global communication frequency
-[no]v bool yes Be loud and noisy
-[no]compact bool yes Write a compact log file
-[no]seppot bool no Write separate V and dVdl terms for each
interaction type and node to the log file(s)
-pforce real -1 Print all forces larger than this (kJ/mol nm)
-[no]reprod bool no Try to avoid optimizations that affect binary
reproducibility
-cpt real 15 Checkpoint interval (minutes)
-[no]cpnum bool no Keep and number checkpoint files
-[no]append bool yes Append to previous output files when continuing
from checkpoint instead of adding the simulation
part number to all file names
-maxh real -1 Terminate after 0.99 times this time (hours)
-multi int 0 Do multiple simulations in parallel
-replex int 0 Attempt replica exchange periodically with this
period (steps)
-reseed int -1 Seed for replica exchange, -1 is generate a seed
-[no]ionize bool no Do a simulation including the effect of an X-Ray
bombardment on your system
Back Off! I just backed up et_an.tpr.log to ./#et_an.tpr.log.2#
Getting Loaded...
Reading file et_an.tpr, VERSION 4.5.5 (single precision)
Loaded with Money
Back Off! I just backed up et_an.tpr.trr to ./#et_an.tpr.trr.2#
Back Off! I just backed up et_an.tpr.edr to ./#et_an.tpr.edr.2#
starting mdrun 'first one'
250000 steps, 250.0 ps.
step 249900, remaining runtime: 0 s
Writing final coordinates.
Back Off! I just backed up et_an.tpr.gro to ./#et_an.tpr.gro.2#
step 250000, remaining runtime: 0 s
NODE (s) Real (s) (%)
Time: 3.760 3.974 94.6
(Mnbf/s) (MFlops) (ns/day) (hour/ns)
Performance: 0.000 363.599 5744.704 0.004
gcq#46: "I Wonder, Should I Get Up..." (J. Lennon)
:-) G R O M A C S (-:
Gyas ROwers Mature At Cryogenic Speed
:-) VERSION 4.5.5 (-:
Written by Emile Apol, Rossen Apostolov, Herman J.C. Berendsen,
Aldert van Buuren, Pär Bjelkmar, Rudi van Drunen, Anton Feenstra,
Gerrit Groenhof, Peter Kasson, Per Larsson, Pieter Meulenhoff,
Teemu Murtola, Szilard Pall, Sander Pronk, Roland Schulz,
Michael Shirts, Alfons Sijbers, Peter Tieleman,
Berk Hess, David van der Spoel, and Erik Lindahl.
Copyright (c) 1991-2000, University of Groningen, The Netherlands.
Copyright (c) 2001-2010, The GROMACS development team at
Uppsala University & The Royal Institute of Technology, Sweden.
check out http://www.gromacs.org for more information.
This program is free software; you can redistribute it and/or
modify it under the terms of the GNU General Public License
as published by the Free Software Foundation; either version 2
of the License, or (at your option) any later version.
:-) mdrun (-:
Option Filename Type Description
------------------------------------------------------------
-s et_sd.tpr Input Run input file: tpr tpb tpa
-o et_sd.tpr.trr Output Full precision trajectory: trr trj cpt
-x et_sd.tpr.xtc Output, Opt. Compressed trajectory (portable xdr format)
-cpi et_sd.tpr.cpt Input, Opt. Checkpoint file
-cpo et_sd.tpr.cpt Output, Opt. Checkpoint file
-c et_sd.tpr.gro Output Structure file: gro g96 pdb etc.
-e et_sd.tpr.edr Output Energy file
-g et_sd.tpr.log Output Log file
-dhdl et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-field et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-table et_sd.tpr.xvg Input, Opt. xvgr/xmgr file
-tablep et_sd.tpr.xvg Input, Opt. xvgr/xmgr file
-tableb et_sd.tpr.xvg Input, Opt. xvgr/xmgr file
-rerun et_sd.tpr.trr Input, Opt. Trajectory: xtc trr trj gro g96 pdb cpt
-tpi et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-tpid et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-ei et_sd.tpr.edi Input, Opt. ED sampling input
-eo et_sd.tpr.edo Output, Opt. ED sampling output
-j et_sd.tpr.gct Input, Opt. General coupling stuff
-jo et_sd.tpr.gct Output, Opt. General coupling stuff
-ffout et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-devout et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-runav et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-px et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-pf et_sd.tpr.xvg Output, Opt. xvgr/xmgr file
-mtx et_sd.tpr.mtx Output, Opt. Hessian matrix
-dn et_sd.tpr.ndx Output, Opt. Index file
-multidir et_sd.tpr Input, Opt., Mult. Run directory
Option Type Value Description
------------------------------------------------------
-[no]h bool no Print help info and quit
-[no]version bool no Print version info and quit
-nice int 0 Set the nicelevel
-deffnm string et_sd.tpr Set the default filename for all file options
-xvg enum xmgrace xvg plot formatting: xmgrace, xmgr or none
-[no]pd bool no Use particle decompostion
-dd vector 0 0 0 Domain decomposition grid, 0 is optimize
-nt int 1 Number of threads to start (0 is guess)
-npme int -1 Number of separate nodes to be used for PME, -1
is guess
-ddorder enum interleave DD node order: interleave, pp_pme or cartesian
-[no]ddcheck bool yes Check for all bonded interactions with DD
-rdd real 0 The maximum distance for bonded interactions with
DD (nm), 0 is determine from initial coordinates
-rcon real 0 Maximum distance for P-LINCS (nm), 0 is estimate
-dlb enum auto Dynamic load balancing (with DD): auto, no or yes
-dds real 0.8 Minimum allowed dlb scaling of the DD cell size
-gcom int -1 Global communication frequency
-[no]v bool yes Be loud and noisy
-[no]compact bool yes Write a compact log file
-[no]seppot bool no Write separate V and dVdl terms for each
interaction type and node to the log file(s)
-pforce real -1 Print all forces larger than this (kJ/mol nm)
-[no]reprod bool no Try to avoid optimizations that affect binary
reproducibility
-cpt real 15 Checkpoint interval (minutes)
-[no]cpnum bool no Keep and number checkpoint files
-[no]append bool yes Append to previous output files when continuing
from checkpoint instead of adding the simulation
part number to all file names
-maxh real -1 Terminate after 0.99 times this time (hours)
-multi int 0 Do multiple simulations in parallel
-replex int 0 Attempt replica exchange periodically with this
period (steps)
-reseed int -1 Seed for replica exchange, -1 is generate a seed
-[no]ionize bool no Do a simulation including the effect of an X-Ray
bombardment on your system
Back Off! I just backed up et_sd.tpr.log to ./#et_sd.tpr.log.2#
Getting Loaded...
Reading file et_sd.tpr, VERSION 4.5.5 (single precision)
Loaded with Money
Back Off! I just backed up et_sd.tpr.trr to ./#et_sd.tpr.trr.2#
Back Off! I just backed up et_sd.tpr.edr to ./#et_sd.tpr.edr.2#
starting mdrun 'first one'
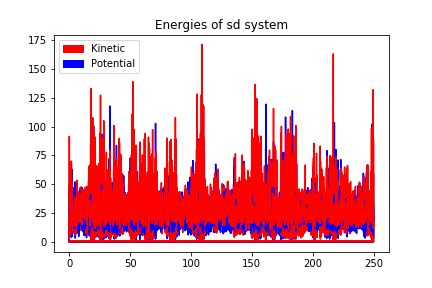
250000 steps, 250.0 ps.
step 249900, remaining runtime: 0 s
Writing final coordinates.
Back Off! I just backed up et_sd.tpr.gro to ./#et_sd.tpr.gro.2#
step 250000, remaining runtime: 0 s
NODE (s) Real (s) (%)
Time: 4.470 4.673 95.7
(Mnbf/s) (MFlops) (ns/day) (hour/ns)
Performance: 0.000 319.124 4832.234 0.005
gcq#255: "This is Tense !" (Star Wars Episode I The Phantom Menace)