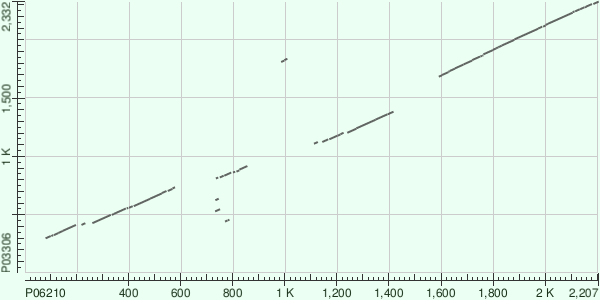
Fig.1 Plot of P06210.3 vs P03306.2

| Identities | Positives | Length, P06210 | Length, P03306 | Gaps | Score | Score (bits) |
|---|---|---|---|---|---|---|
| 29% | 48% | 651 | 644 | 59 | 636 | 249 |
| 36% | 49% | 294 | 263 | 43 | 397 | 157 |
Alignment 1) The aligned part of P06210 is entirely in Protein 3CD; of P03306 - in Picornain 3C and RNA-directed RNA polymerase 3D-POL. Alignment 2) The aligned part of P06210 is mostly in Protein 2C; of P03306 - entirely in Protein 2C.
| ID1 | ID2 | Score | Median | Quartile | Score (bits) | Probability | |
|---|---|---|---|---|---|---|---|
| Homologic | CLPP_BACSU | CLPP_ECOLI | 695.0 | 37 | 41.75 | 139.5 | 1e-42 |
| Non-homologic | CLPP_BACSU | SYC_ECOLI | 58.0 | 39 | 44.75 | 4.3 | 5e-2 |
| Identities | Positives | Length, ALV23621.1 | Length, second | Gaps | Score | Score (bits) | Expect | % Coverage | |
|---|---|---|---|---|---|---|---|---|---|
| Q9ZLX4.1 | 39% | 58% | 356 | 374 | 18 | 616 | 241 | 5e-76 | 100% |
| O25242.1 | 39% | 58% | 356 | 374 | 18 | 596 | 234 | 5e-73 | 100% |