3. A- and В- DNA forms. RNA structure
Task 1
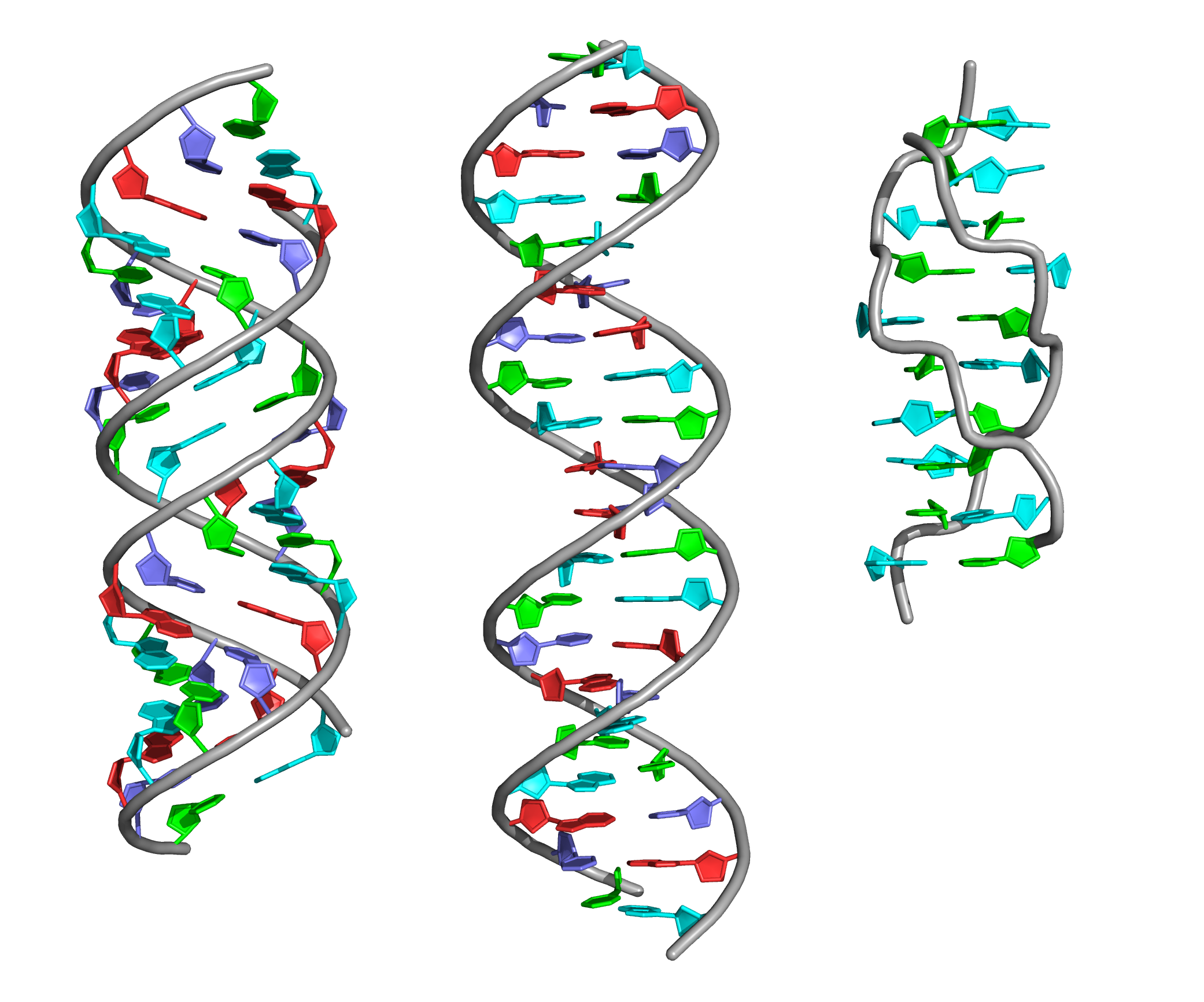
Using fiber program from 3DNA package, 3 structual forms of the DNA sequence GATC (repeated 5 times) were obtained. Download .pdb files: A-NDA form, B-DNA form , Z-DNA form .
Task 2
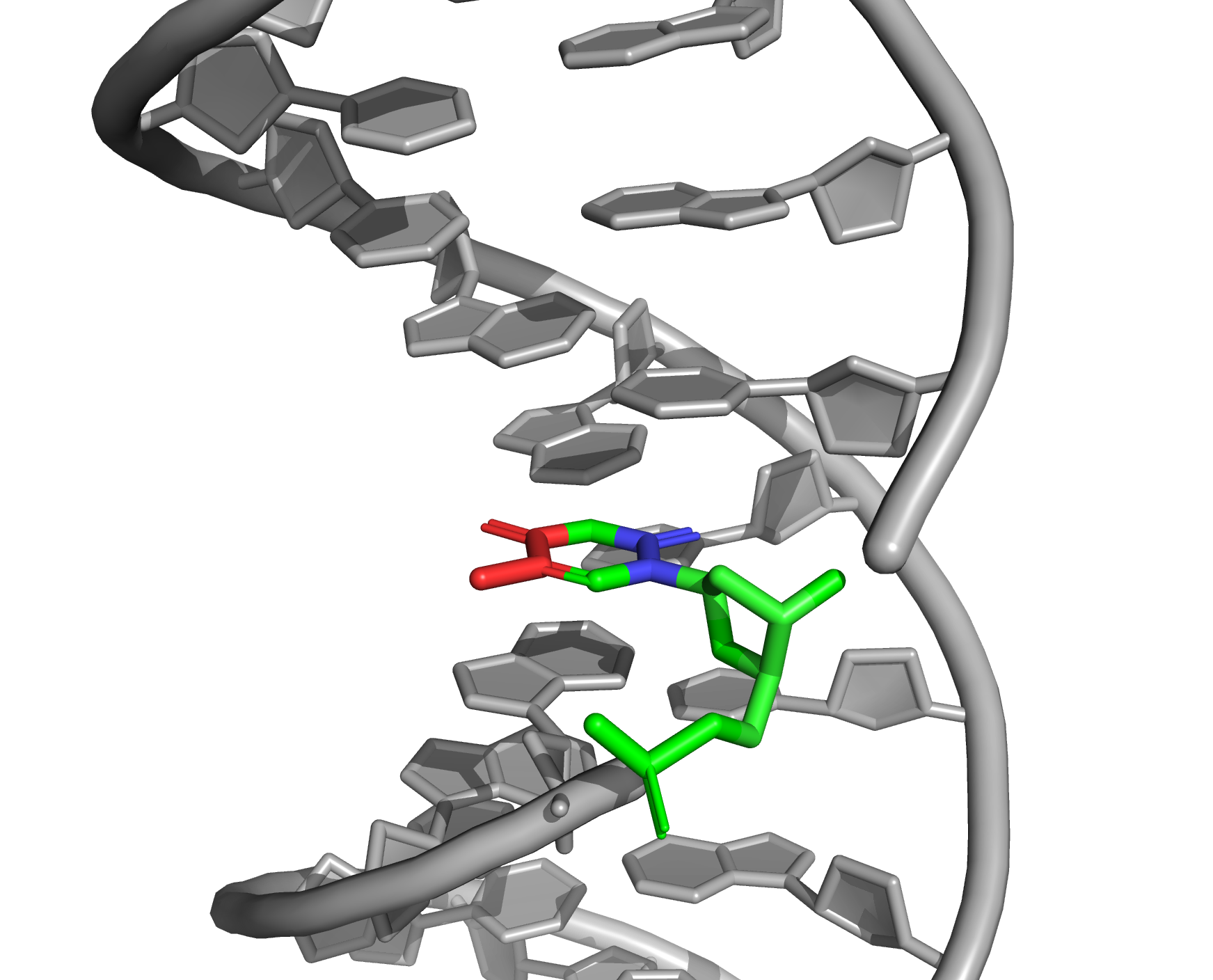
Let's look at the B-DNA from and the location of certain thymine in space (Fig.3). It was determined which atoms are directed towards the major or minor groove (Fig.2). Towards the major groove: C7, C5, C4, O4. Towards the minor groove: O2, C2, N1.
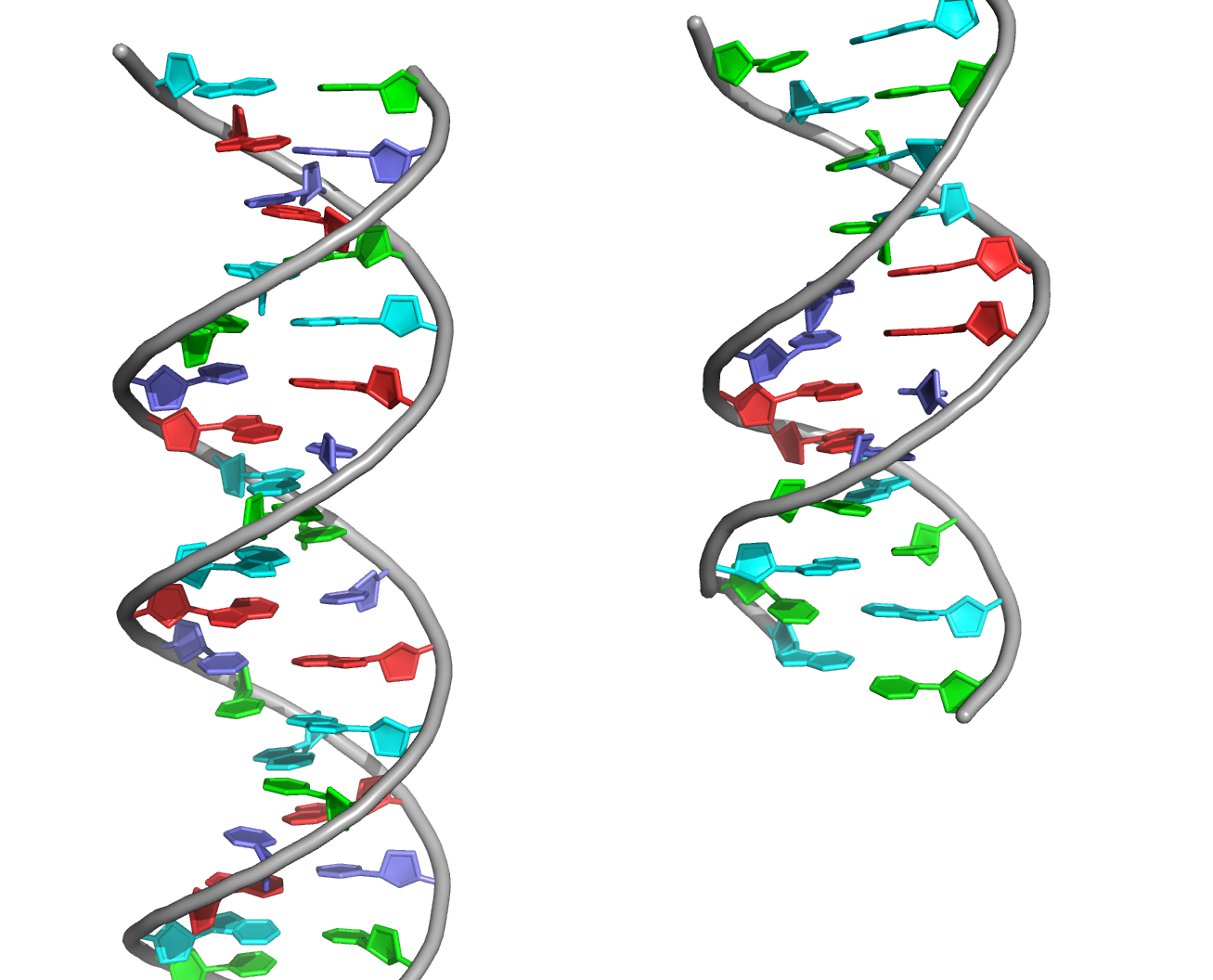
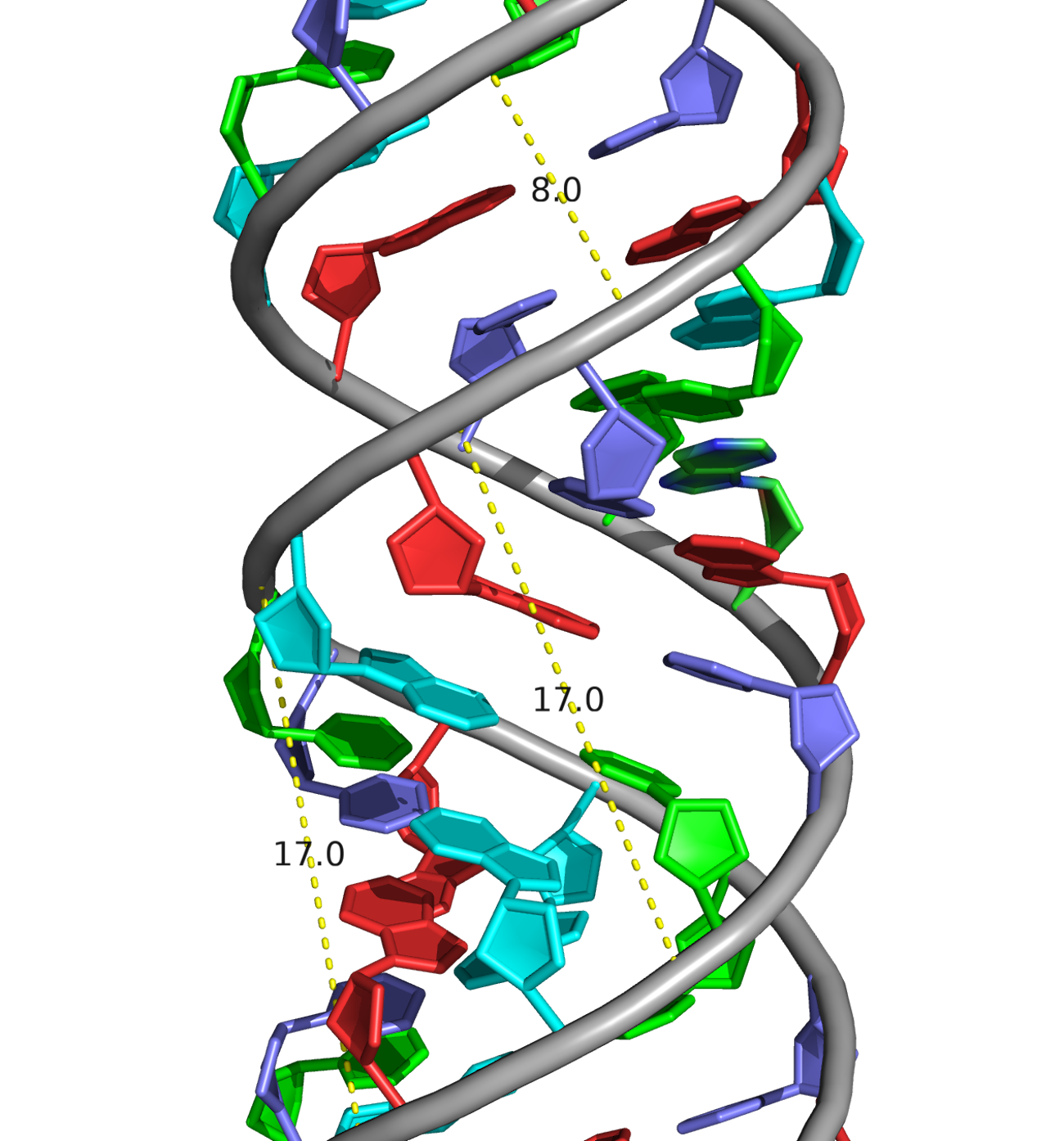
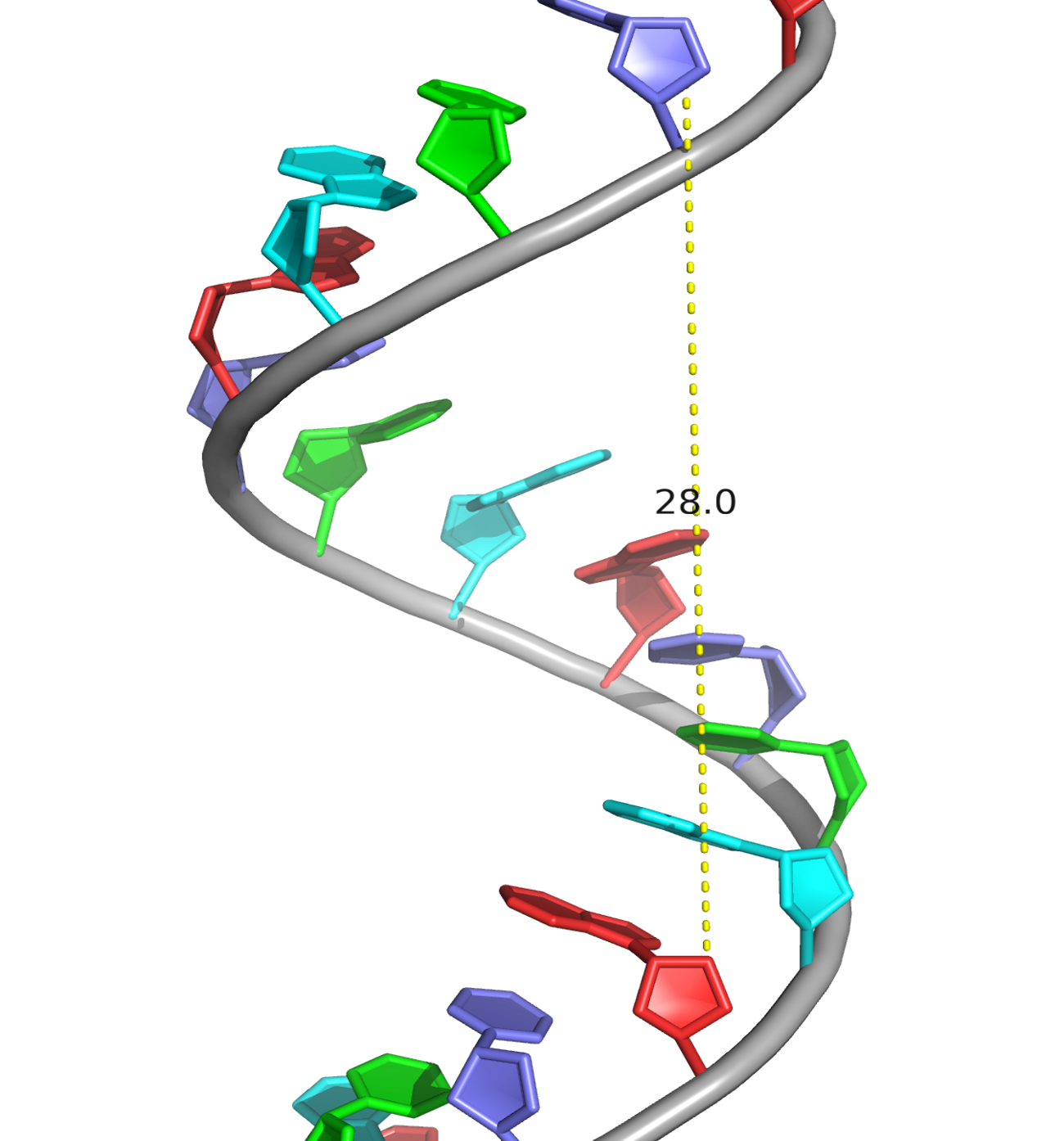
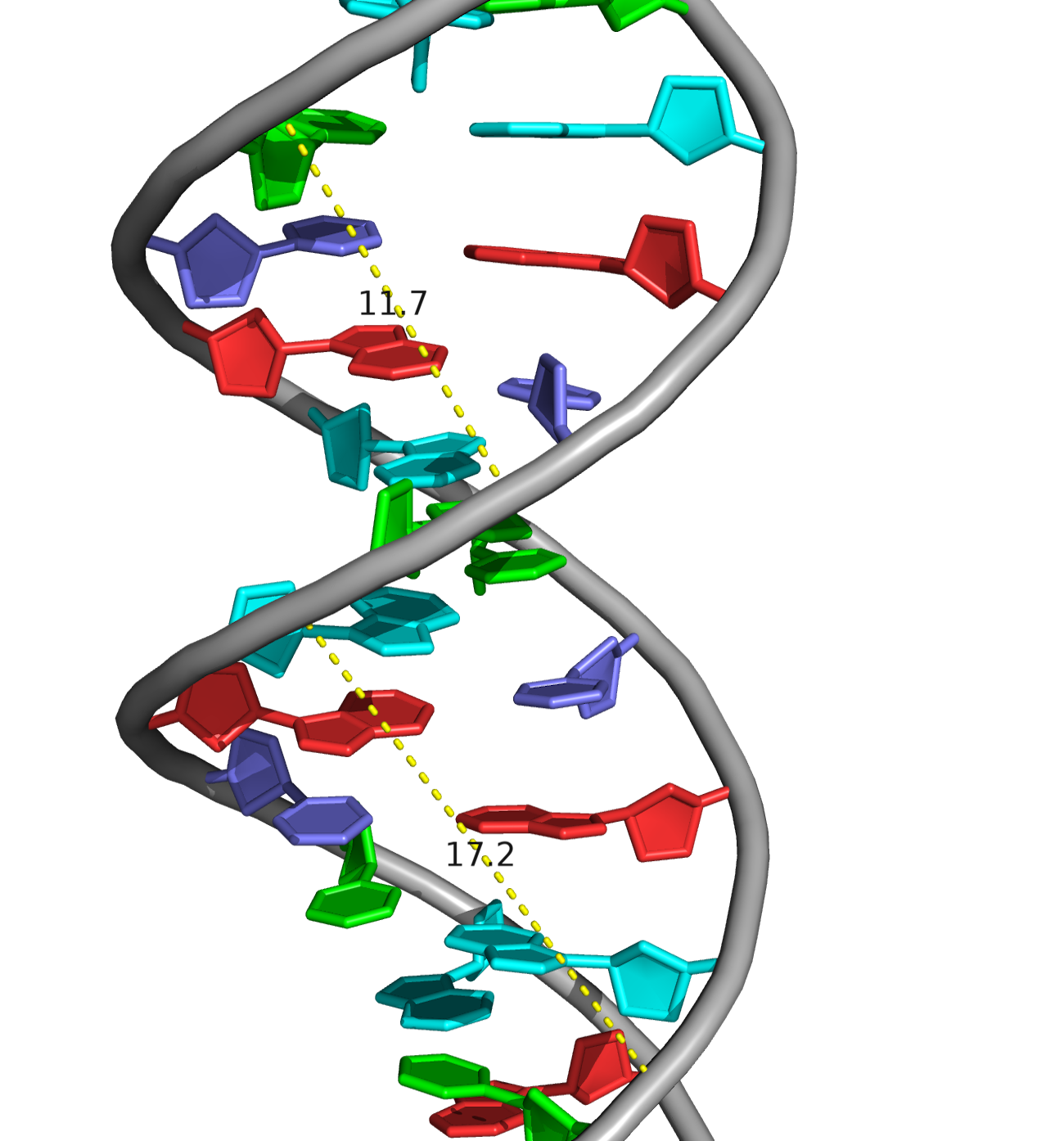
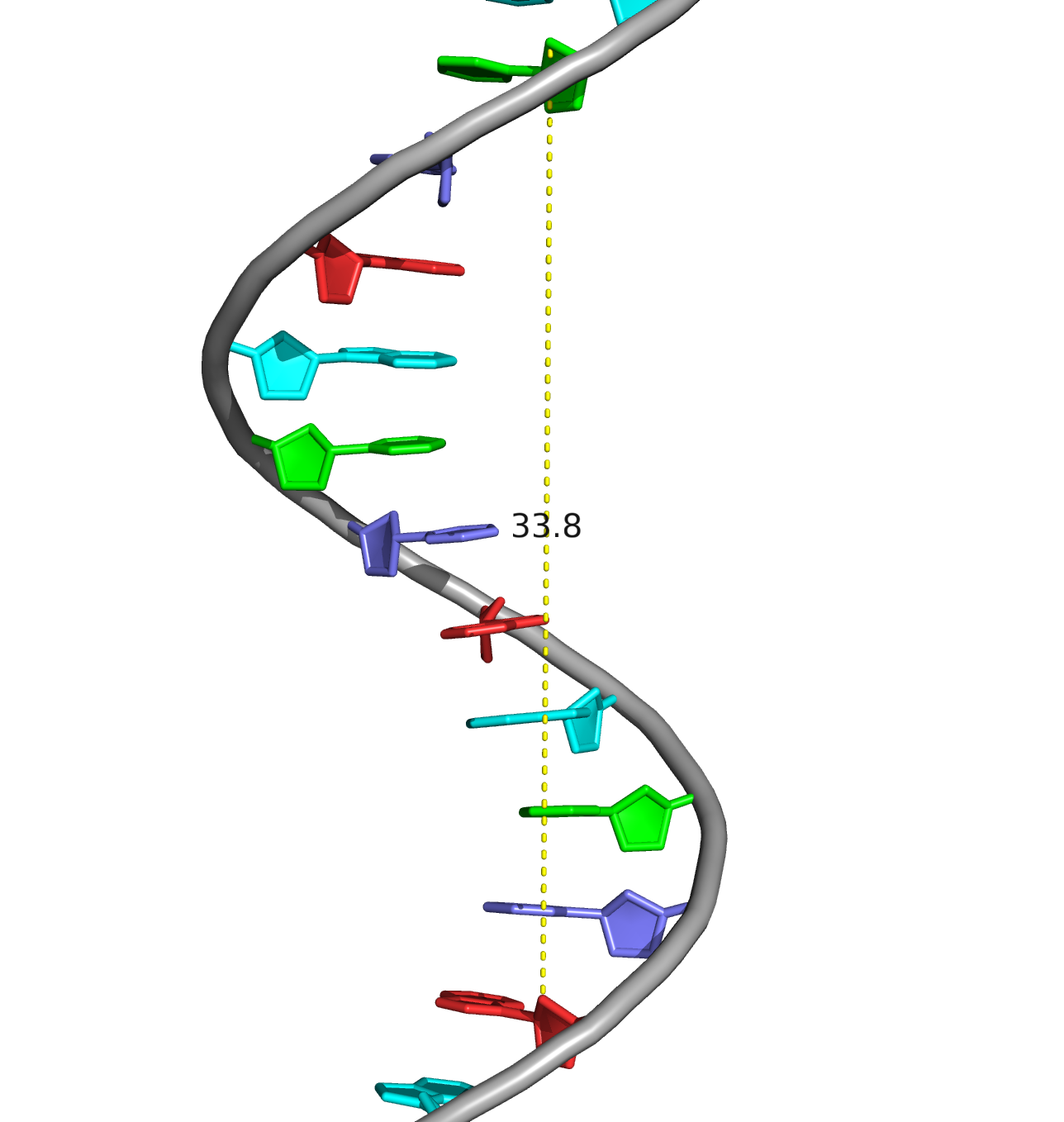
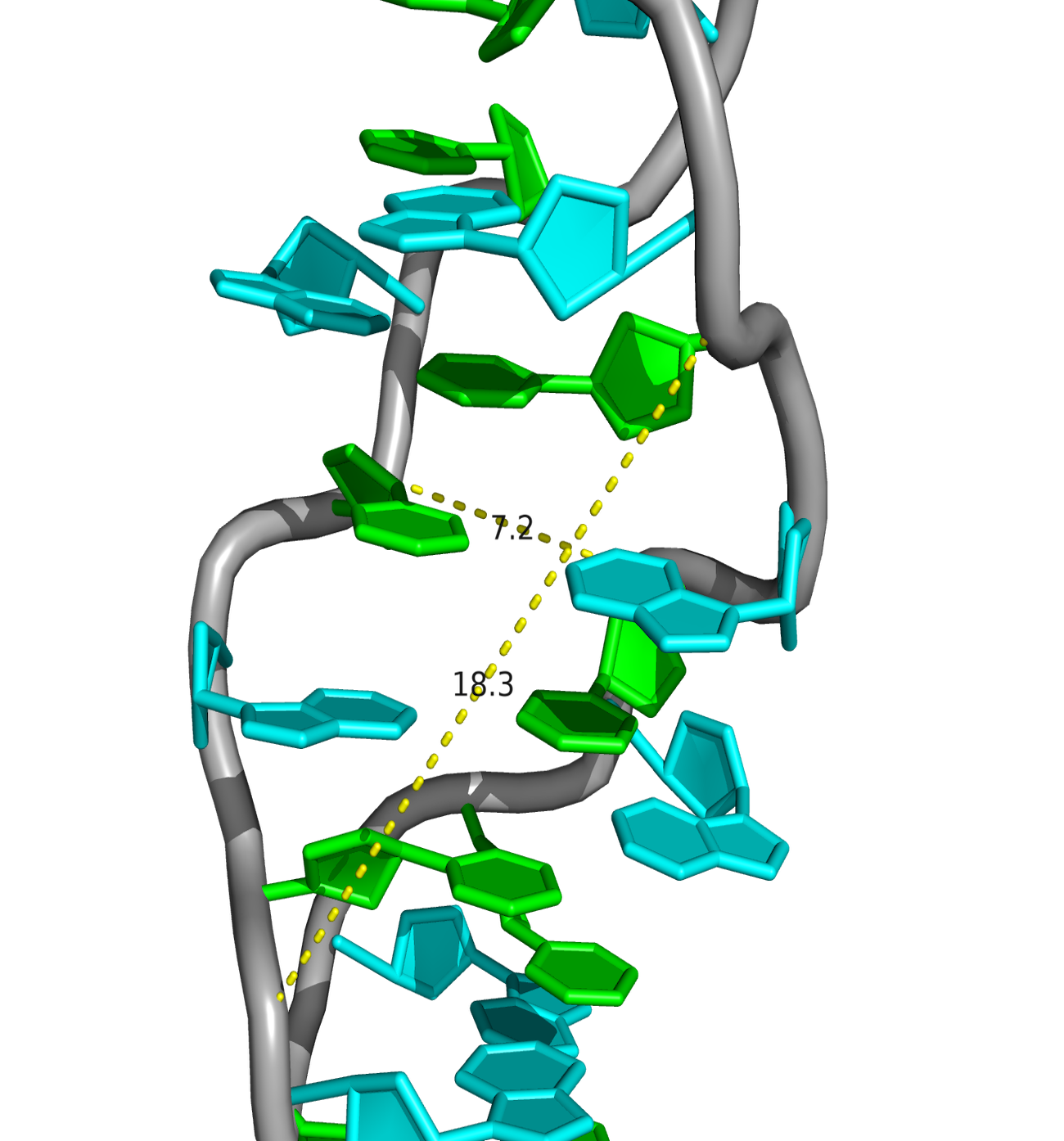
A visual comparison of the experimental DNA-B form and DNA-B from task 1 was made (Fig.4) The helix pitch (Å), the nuber of nucleotides per turn, and the widths of the major and minor group were also found for each DNA from (Fig.5-9). The results are present in the table below (Table 1):
| A-DNA from | B-DNA from | Z-DNA from | |
|---|---|---|---|
| Spiral type | Right | Right | Left |
| Spiral pitch (Å) | 28.0 | 33.8 | 43.5 |
| Number of bases per turn | 11 | 10 | 12 |
| Width of major groove (Å) | 17.0 (A/DA`34-B/DG`9) | 17.2 (A/DA`34-B/DC`4) | 18.3 (A/DC`4-B/DC`14) |
| Width of minor groove (Å) | 8.0 (A/DA`12-B/DG`9) | 11.7 (A/DA`26-B/DG`9) | 7.2 (A/DG`31-B/DG`13) |