Standart protein as in most of exercises was used PDB ID 4ri1. Below is the comparison of the secondary structures found by stride -o 4ri1.pdb and described in the original pdb file. In both files residues are numbered the same making possible direct comparison. Protein of interest with coloring by ss is on the fig.1.
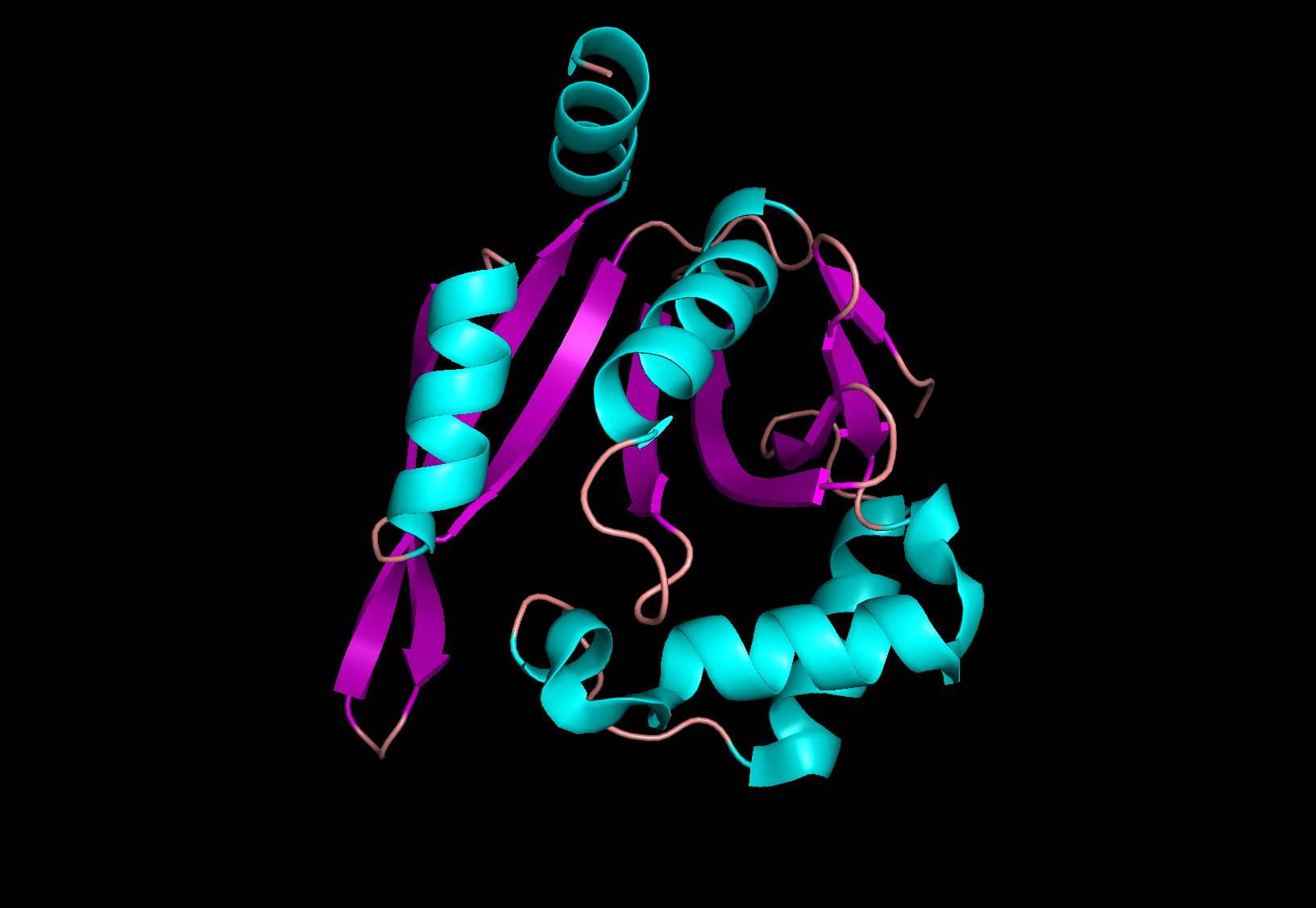
Figure 1. protein 4ri1 colored by ss
HELIX 5 5 GLY A 102 GLU A 116 1 15 HELIX 6 6 ASN A 131 ASN A 141 1 11 SHEET 1 A 8 TYR A 5 TYR A 7 0 SHEET 2 A 8 ILE A 10 ASP A 14 -1 O ALA A 12 N TYR A 5 SHEET 3 A 8 ARG A 63 GLU A 69 -1 O LEU A 66 N ILE A 13 SHEET 4 A 8 VAL A 72 ASN A 83 -1 O GLY A 75 N PHE A 67 SHEET 5 A 8 HIS A 88 LYS A 95 -1 O TYR A 94 N VAL A 76 SHEET 6 A 8 SER A 122 MET A 128 1 O HIS A 124 N GLY A 89 SHEET 7 A 8 GLU A 158 ASP A 168 -1 O LEU A 163 N VAL A 127 SHEET 8 A 8 GLU A 144 LYS A 155 -1 N LYS A 155 O GLU A 158
LOC AlphaHelix GLY 103 A GLU 116 A 4RI1 LOC AlphaHelix PHE 132 A ASN 141 A 4RI1 LOC Strand TYR 5 A TYR 7 A 4RI1 LOC Strand ILE 10 A ASP 14 A 4RI1 LOC Strand ARG 63 A GLU 69 A 4RI1 LOC Strand VAL 72 A ASN 83 A 4RI1 LOC Strand HIS 88 A LYS 95 A 4RI1 LOC Strand SER 122 A MET 128 A 4RI1 LOC Strand GLU 144 A LYS 155 A 4RI1 LOC Strand GLU 158 A ASP 168 A 4RI1
As it can be seen all beta-strands were predicted precisely by stride, and alpha-helixes were both off by one at the beginning.
Term 7 Main page