Structures of Staphylococcal Nuclease were reviewed: 2LKV (NMR) and 1A3T (X-Ray). NMR structure has 10 models; X-Ray structure has a resolution of 2.1 angstroms. Even though their structures' images don't look alike (on the website), they are quite similar (Fig. 1).
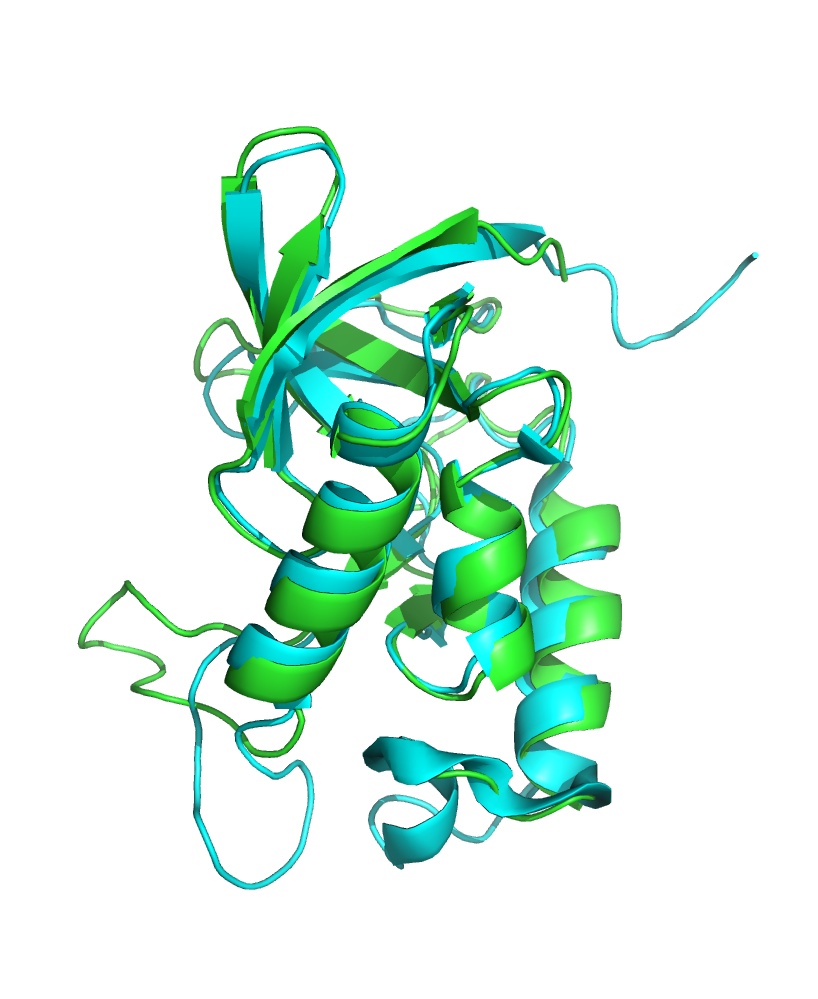
Figure 1. Superposition of the structures.
1. The selected backbone hydrogen bond in the core of the protein was between ASN-100/O and VAL-104/N atoms. Both residues are part of an alpha-helix and are located inside the core.
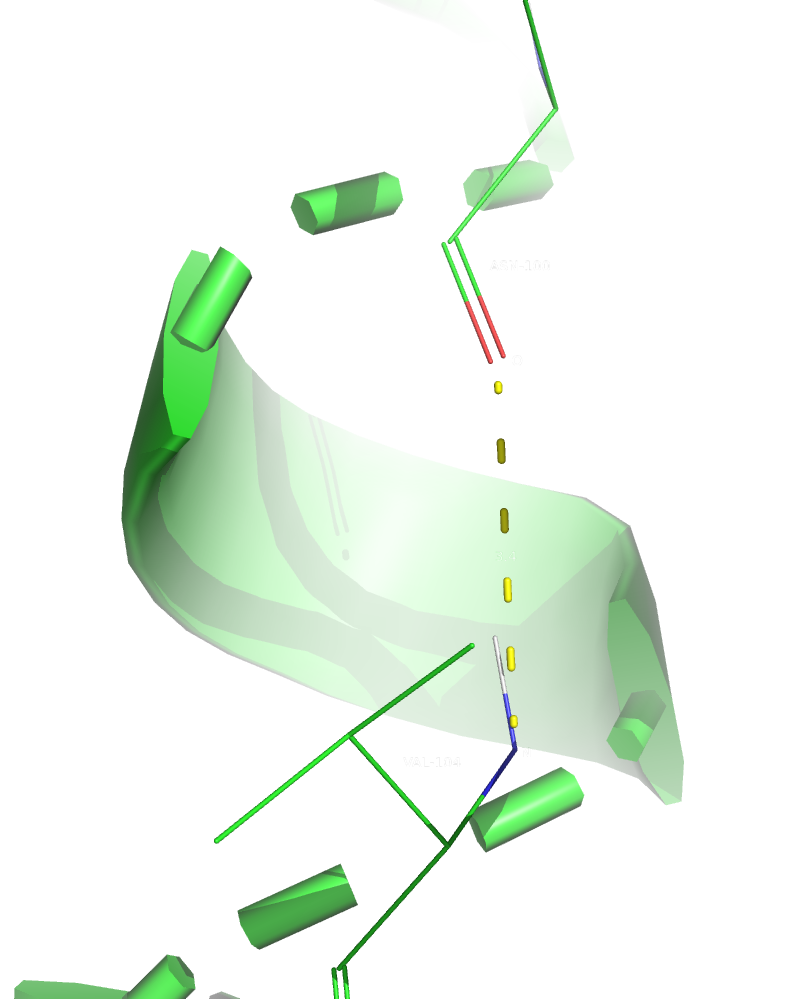
Figure 2. The first hydrogen bond (in X-Ray structure).
2. I couldn't find a hydrogen bond between two sidechains in the core, so I decided to select a sidechain-backbone one. The selected hydrogen bond was between ALA-90/O and ASN-100/ND2 atoms. ALA-90 is a part of a beta-sheet and ASN-100 is a a part of an alpha-helix. Both residues are located inside the core.
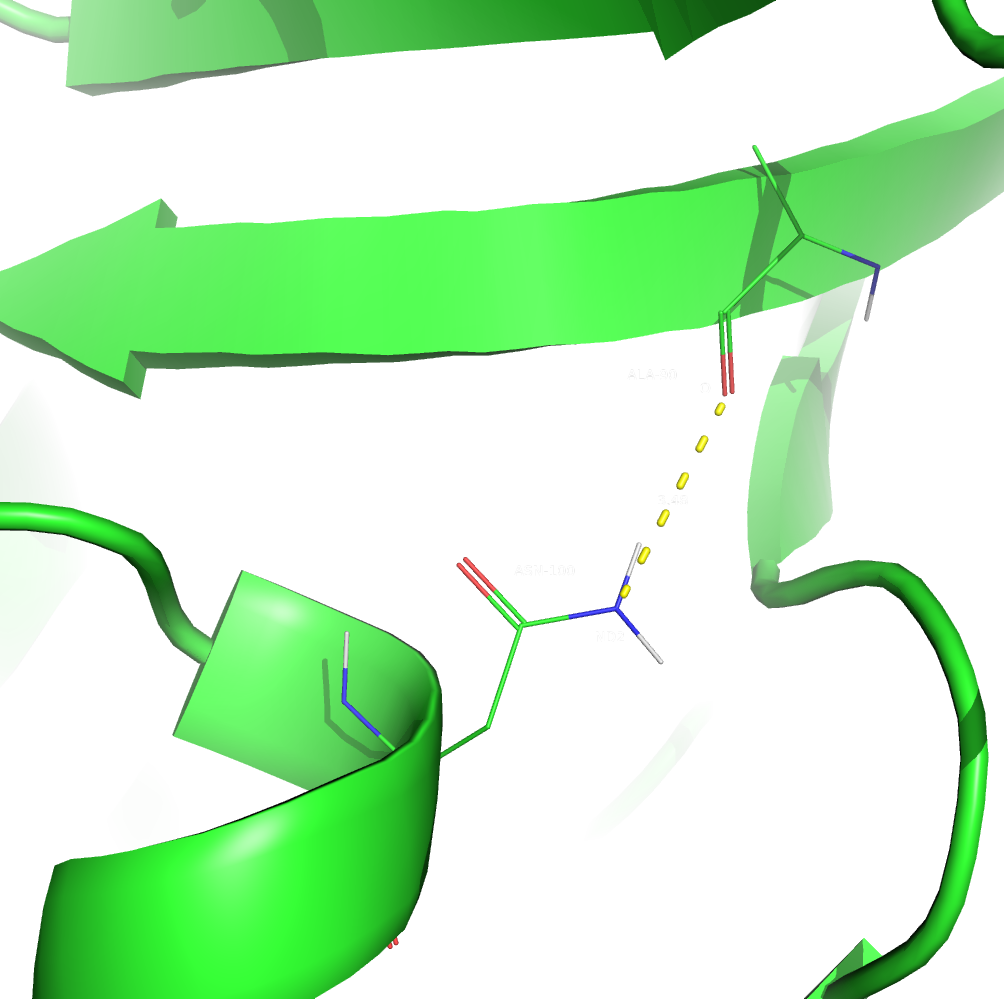
Figure 3. The second hydrogen bond (in X-Ray structure).
3. The selected hydrogen bond in a loop on the surface of the protein was between GLU-43/O and GLU-52/N atoms. Both residues are part of a loop and are located on the surface. It is also worth mentioning that they form one additional hydrogen bond (GLU-52's sidechain oxygen with GLU-43's backbone nitrogen).
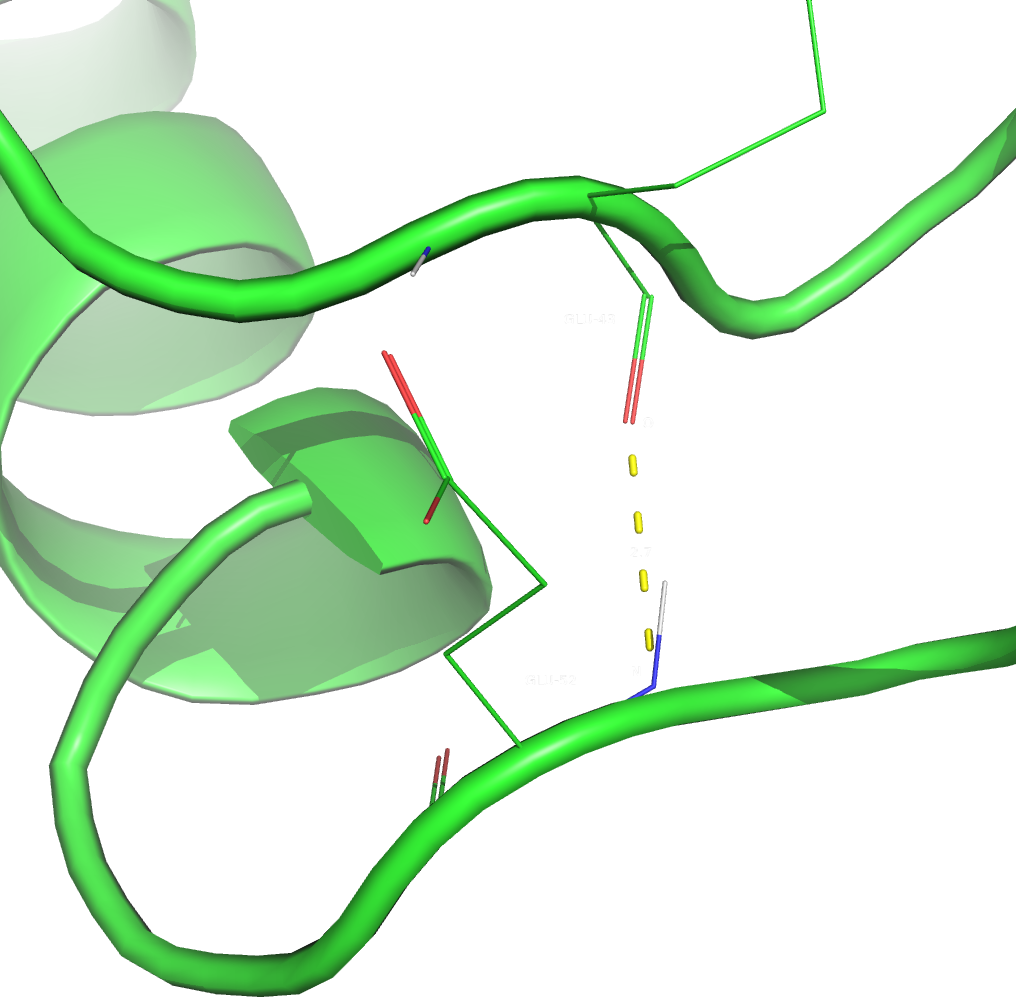
Figure 4. The third hydrogen bond (in X-Ray structure).
Table 1. Comparison of selected hydrogen bonds in X-Ray and NMR structures.
| Hydrogen bond ID | Distance in X-Ray | Number (per cent) of NMR models with the bond | Min., max. and median distance in NMR |
|---|---|---|---|
| 1 | 3.4 | 10 (100%) | 2.8, 2.9, 2.9 |
| 2 | 3.48 | 0 (0%) | 3.52, 4.04, 3.725 |
| 3 | 2.7 | 0 (0%) | 5.6, 9.7, 7.05 |
In my opinion, no pictures are necessary to draw conclusions from the Table 1. The first hydrogen bond (which was between backbone atoms in an alpha-helix inside the core) remained in all NMR models. It's length, however, notably decreased with almost no fluctuations among the models. This could mean that the alpha-helix is less streched and is more rigid when the protein is in the solution. The other two bonds weren't observed at all in NMR models, so they probably form almost only upon crystallization, which makes sense as the first bond is too weak even in a crystall (it's length is almost 3.5 angstroms) and the second one is in a loop, and loops are known to be quite flexible even in crystalls.
Term 7 Main page