A protein with PDB ID 4RI1, chain A, was used (same as in the Task 1). Two structure homologs were chosen: 4R87, 3WR7, -- chains F and D respectively; RMSDs of 1.57 and 1.61.
Structural alignment (FASTA) Structures' superposition (PDB) JalView project with alignments (jvp)For comparison, a sequential only alignment was costructed using MUSCLE service. Two differently aligned residues were chosen (highlighted with red rectangles on Fig. 1). The two bottom proteins are aligned similarly, but the first one is aligned differently.
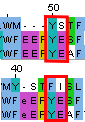
Figure 1. Alignment regions of interest (MUSCLE on top; PDBeFold on bottom).
To determine which falignment is correct (for the selected region) structures' superposition was analyzed.
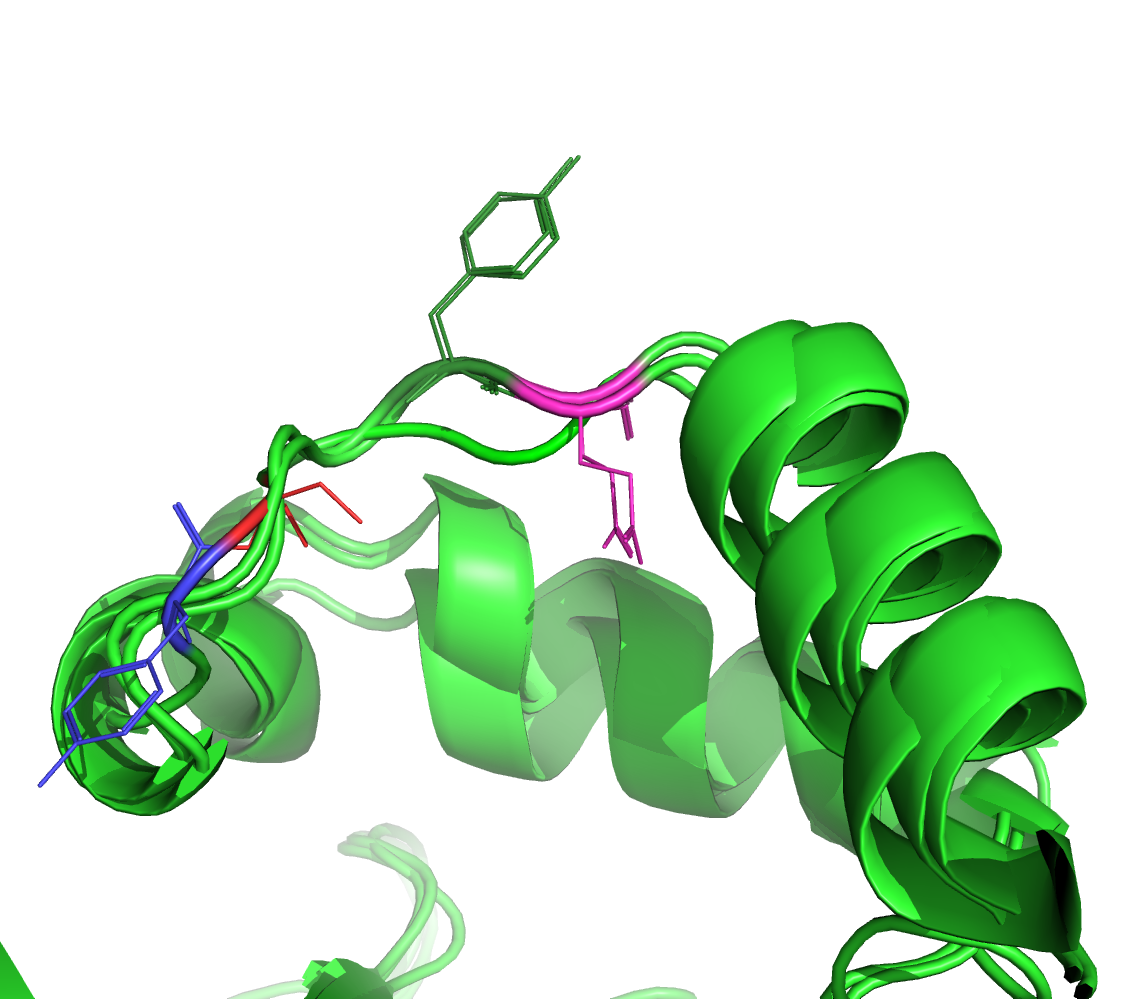
Figure 2. Alignment region of interest (MUSCLE); aligned residues are color coded -- dark green/magenta for the bottom two proteins and red/blue for the top one.
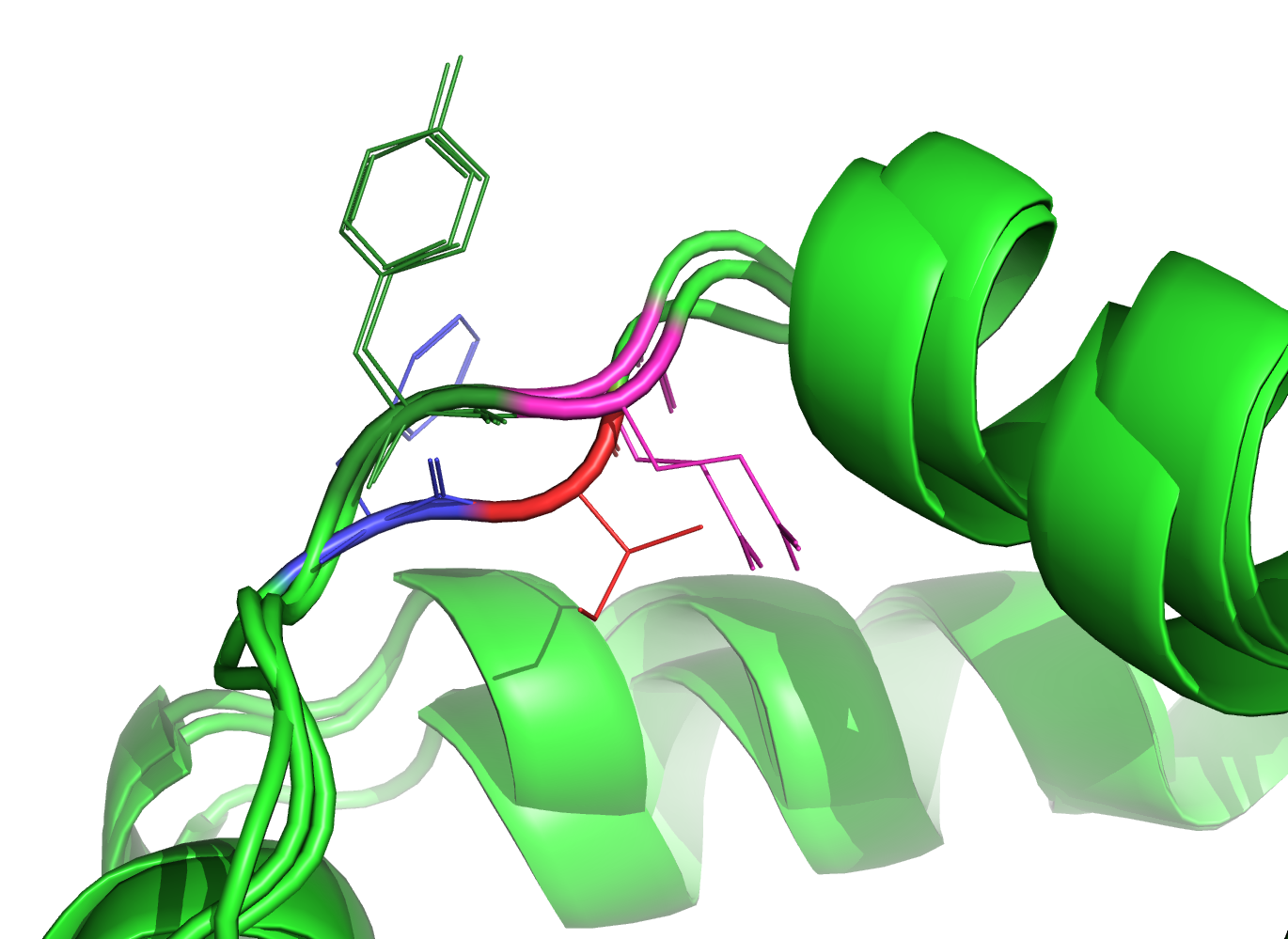
Figure 3. Alignment region of interest (PDBeFold); aligned residues are color coded -- dark green/magenta for the bottom two proteins and red/blue for the top one.
Figures 2-3 suggest that the structural alignment aligned the inspected residues better (according to the superposition of the structures).
Term 7 Main page