The local-resemblance map of two polyproteins
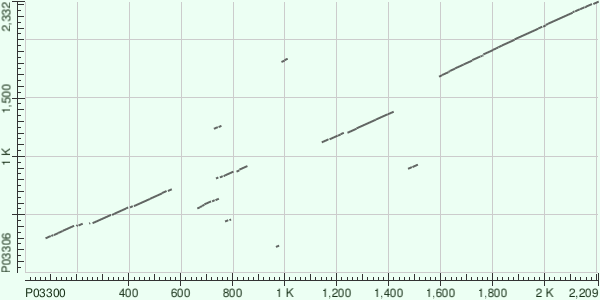
|
|
The best Score results are 250 and 157 bits.
For the first result:
PO330 - Protein 3CD
P03306 - RNA-directed RNA polymerase 3D-POL (1863 - 2332), Picornain 3C (1650 (1684) - 1862;
probably, there are one protein and the piece of other protein in this alignment
For the second result:
P0330 - Protein 2C (probably, the part of rhe protein)
Po3306 - Protein 2C (probably, the part of rhe protein)
Comparing the weight of alignment with random
| ID1, ID2 | Score | Median (m) | Upper quartile (Q1) | Bits (B) | Probability (P) |
| TRPB_ECOLI, TRPB_BACSU | 1102.0 | 55.25 | 62.25 | 149.7 | 8.626E-46 |
| DNAA_ECOLI, FADA_ECOLI | 55.5 | 45.0 | 50.0 | 2.3 | 0.203 |
So, we can see that the first alignment is not accidentally; and the second may be appeared because of the occasional reasons
Increasing of penalty for gap extension (= 4.0)
| ID1, ID2 | Score | Median (m) | Upper quartile (Q1) | Bits (B) | Probability (P) |
| TRPB_ECOLI, TRPB_BACSU | 1100 | 36 | 38.00 | 532.5 | 4.03E-161 |
| DNAA_ECOLI, FADA_ECOLI | 44 | 34 | 37 | 3.7 | 0.077 |
So, we can notice that increasing of penalty for gap extension (= 4.0)
gives us decrease of the probability of accidental alignment
Alignments using BLOSUM30
| ID1, ID2 | Score | Median (m) | Upper quartile (Q1) | Bits (B) | Probability (P) |
| TRPB_ECOLI, TRPB_BACSU | 1498 | 297.75 | 308.75 | 109.2 | 1.34E-33 |
| DNAA_ECOLI, FADA_ECOLI | 264 | 277.5 | 289.25 | -1.06 (uncorrect) | 2.08 (uncorrect) |
With the help of BLOSUM30 we got results with higher significans of probability (P).
We can also see negative bit values, which are incorrect (even with the recount of score).
Thus, we can say that BLOSUM30 does not always work properly
Checking the formula for conversion to bits
Let's check our results using standard parameters.
In the first case we will look at trpb_ecoli and trpb_bacsu
and in the second case - dnaa_ecoli and fada_ecoli.
We will shuffle trp_bacsu and fada_ecoli sequences for 1000 terms
| ID1, ID2 | Upper Quartile (Q1) | Median (m) | Upper 1/8 | Bits (B) | Probability (P) |
| TRPB_ECOLI, TRPB_BACSU | 61.25 | 54.5 | 67.5 | 2.1 | 0.233 |
| DNAA_ECOLI, FADA_ECOLI | 50.5 | 45.0 | 55.0 | 2 | 0.25 |
The bits'results smaller than 3 for a while
BLAST: search for homologs in the database
| AC | ID | Organism | Identities, % | Positives, % | Length | Gaps | Score | Score, bits | Expect | Coverage, % |
| Q49492 | MYCF_MICGR | Micromonospora griseorubida | 42 | 58 | 281, 253 | 34 | 602 | 236 | 1,00E-76 | 100 |
| Q9L9F2 | NOVP_STRNV | Streptomyces niveus (Streptomyces spheroides) | 51 | 72 | 207, 209 | 2 | 592 | 232 | 5,00E-75 | 73,7 |