|
Organism (taxonomy): |
||||||||||||||||||
Some comments for Table1
There're 2 PDB records according to the Table 1 (3N3U, 4ITR).
HMW IgBP has two chains - Chain1 (98 – 4095; Adenosine monophosphate-protein transferase and cysteine protease IbpA)
and Chain2 (3222 – 4095 ; Protein p76 IgBP).
Also there's some information about Signal peptide (1 – 97; it situates before the Chain1). The structure of the whole protein is known.
It is interesting that one chain, as it was written before: 98-4095, another: 3222-4095. Their sequences do not coincide (with the aid of the Python).
This probably means that different forms of the protein product, long and short (different splicing variants), can result.
Protein Existence: evidence at protein level (that's mean that this protein has sufficiently accurate structure).
About the function (from this page):
"... Adenylyltransferase involved in virulence by mediating the addition of adenosine 5'-monophosphate (AMP)
to specific tyrosine residue of host Rho GTPases RhoA, Rac and Cdc42.
The resulting AMPylation inactivates Rho GTPases, thereby inhibiting actin assembly in infected cells.
Probably also acts as a cysteine protease, which may play a central role after invasion of host cell and in virulence.
Possible member (with IbpB) of a 2 partner secretion. Probably able to bind bovine epithelial cells (host cells).
May participate in the formation of fibrils at the surface of the bacteria."
For the first time in the database the protein was entered on August 29, 2003
| Table 2. Information about clusters | ||||
| UniRef | Cluster ID | Size | Organisms | Cluster name |
| UniRef50 | UniRef50_Q06277 | 3 | Histophilus somni (strain 2336) (Haemophilus somnus) | Adenosine monophosphate-protein transferase and cysteine protease IbpA |
| UniRef90 | UniRef90_Q06277 | 2 | Histophilus somni (strain 2336) (Haemophilus somnus) | Adenosine monophosphate-protein transferase and cysteine protease IbpA |
| UniRef100 | UniRef100_Q06277 | 2 | Histophilus somni (strain 2336) (Haemophilus somnus) | Adenosine monophosphate-protein transferase and cysteine protease IbpA |
Sessions of search in the UniProt. Searh by Short or Full name
Search by short nameWe input:name:"hmw igbp" Reviewed (Swiss-Prot): 1 Unreviewed (TrEMBL): 1 Total (number of proteins): 2 |
Search by short name AND organismWe input:hmw igbp organism:histophilus somni (strain 2336) Reviewed (Swiss-Prot): 1 Total (number of proteins): 1 |
Search by short name AND familyWe input:name:"hmw igbp" taxonomy:pasteurellaceae Reviewed (Swiss-Prot): 1 Total (number of proteins): 1 |
Search by short name AND phylumWe input:name:"hmw igbp" taxonomy:proteobacteria Reviewed (Swiss-Prot): 1 Unreviewed (TrEMBL): 1 Total (number of proteins): 2 |
Search by full nameWe input:name:"adenosine monophosphate protein transferase and cysteine protease ibpa" Reviewed (Swiss-Prot): 1 Unreviewed (TrEMBL): 38 Total (number of proteins): 39 |
Search by full name AND organismWe input:name:"adenosine monophosphate protein transferase and cysteine protease ibpa" organism:"histophilus somni strain 2336" Reviewed (Swiss-Prot): 1 Total (number of proteins): 1 |
Search by full name AND familyWe input:name:"adenosine monophosphate protein transferase and cysteine protease ibpa" taxonomy:pasteurellaceae Reviewed (Swiss-Prot): 1 Total (number of proteins): 1 |
Search by full name AND phylumWe input:name:"adenosine monophosphate protein transferase and cysteine protease ibpa" taxonomy:proteobacteria Reviewed (Swiss-Prot): 1 Unreviewed (TrEMBL): 13 Total (number of proteins): 14 |
Search by the word "allergen"
Search by the word "allergen" (speaking about protein allergens)We input:name:allergen Reviewed (Swiss-Prot): 474* Unreviewed (TrEMBL): 9792 Total (number of proteins): 10266 |
Search by protein allergen (Fungi, kingdom)We input:name:allergen taxonomy:fungi Reviewed (Swiss-Prot): 45 (9.49% of 474*) Unreviewed (TrEMBL): 1537 Total (number of proteins): 1582 |
Search by protein allergen (Metazoa, kingdom)We input:name:allergen taxonomy:metazoa Reviewed (Swiss-Prot): 122** (25.74% of 474*) Unreviewed (TrEMBL): 1688 Total (number of proteins): 1810 |
Search by protein allergen (Virdiplantae, kingdom)We input:name:allergen taxonomy:metazoa Reviewed (Swiss-Prot): 307 (64.77% of 474*) Unreviewed (TrEMBL): 1394 Total (number of proteins): 1701 |
Search by protein allergen (Vertebrata)We input:name:allergen taxonomy:vertebrata Reviewed (Swiss-Prot): 22 (18.03% of 122**) Unreviewed (TrEMBL): 38 Total (number of proteins): 60 |
Search by protein allergen (Arthropoda)We input:name:allergen taxonomy:arthropoda Reviewed (Swiss-Prot): 93 (76.23% of 122**) Unreviewed (TrEMBL): 1404 Total (number of proteins): 1497 |
Discussion of some search results
 |
 |
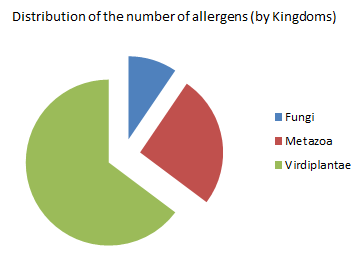
Here you can see two diagrams which was built according to the search results. From the first pie - chart you can see, that the greatest number of allergens (in the UniProt database) come from Virdiplantae kingdom; the least number of allergens at Fungi kingdom. It's interesting that the total number of all allergens in the UniProt (474) consists of "allergenic" representatives from this three kingdoms.
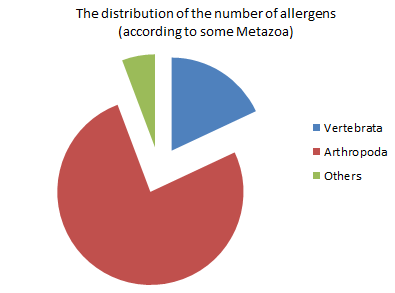
At the second pie chart showed that Arthropoda has the biggest number of allergens comparative to Vertebrata
Search by the word "trypsin"
Search by the word "trypsin"We input:name:trypsin Reviewed (Swiss-Prot): 311*** Unreviewed (TrEMBL): 18640 Total (number of proteins): 18951 |
Search by protein allergen (Fungi, kingdom)We input:name:trypsin annotation:(type:function "trypsin inhibitor") Reviewed (Swiss-Prot): 22 (7.07% of 311***) Total (number of proteins): 22 |
Comparison of RefSeq and UniProt
According to the data from UniProt, the full name of our protein is
"Adenosine monophosphate-protein transferase and cysteine protease IbpA".
RefSeq (WP_012340627.1) gives us the structure definited as "cysteine protease [Histophilus somni]".
In the RefSeq nothing is said about transferase, which was mentioned in the full name of UniProt.
The length of the protein sequence is 4095 amino acid residues both in the RefSeq and the UniProt.
It is important to mention the date of the latest modifications of the records.
In the UniProt we can see 28-FEB-2018 (entry version 105), and in the RefSeq Protein - 14-JUN-2015.
The version in the UniProt is more up to date.
In general, the databases' information is the same.
In my opinion, in RefSeq Protein the data format is more comfort for visual perception.
History of record's changes in UniProt
There're 105 versions of the record of our protein in the History.
The first version was integrated into UniProtKB / Swiss-Prot on August 29, 2003.
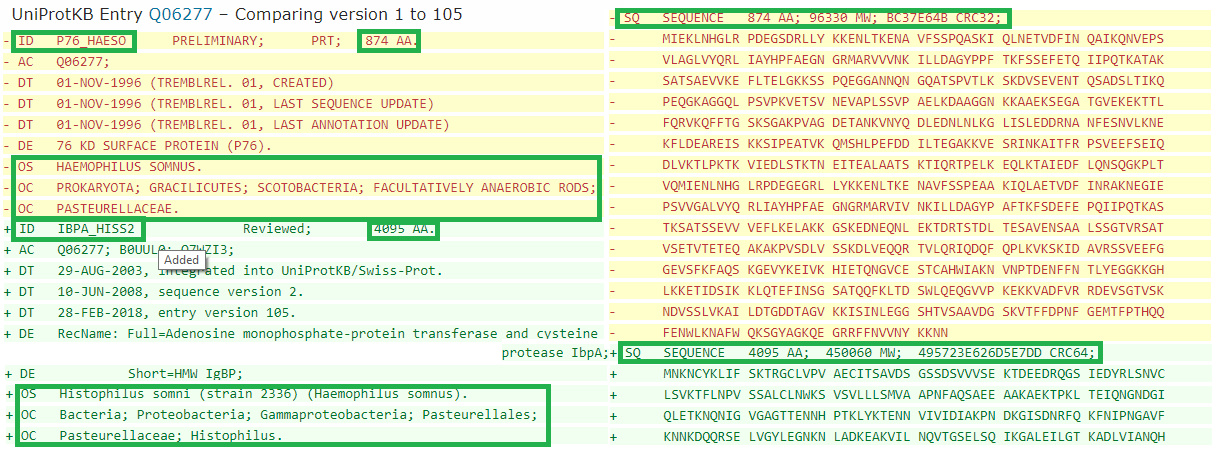
105 version has changed significantly compared to 1. Some noticeable changes are shown in the picture.
Let's describe some changes:
1) The ID was changed (P76_HAESO - IBPA_HISS2)
2) The number of amino acid residues in the structure (874 - 4095)
3) The sample of identifiers became wider (DR)
4) The lines OS and OC have been changed (we can notice the transition from obsolete taxa)
5) In version 105, another amino acid sequence (SQ)
The change from 36 to 37 characterizes by changing of the structure's ID (I've wrote about it earlier).
After 81 version, the structure's ID was unchanged. Feature Table (FT) was added
We can understand the system of UniProt with the help of this page
From all parameters I managed to find some strings about posttranslational modification (PTM, with the help of "Help"). Here they're: The long form of the protein is probably processed, and/or the transcript may be subject to differential translational initiation