MAIN
TERMS
PROJECTS
ABOUT
Alignments
Local similarity map
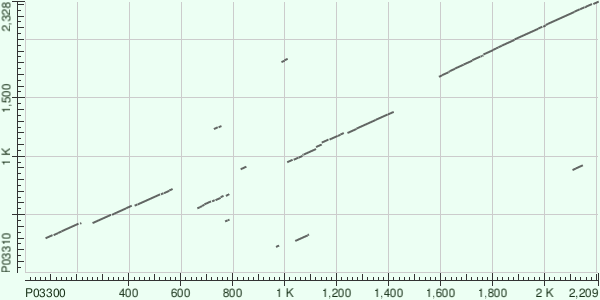
Alignment map

Most relevant alignment
| Identities(%) | Positives(%) | Length(Poliovirus type 1 (strain Mahoney)) | Length(Foot-and-mouth disease virus (isolate -/Brazil/C3Indaial/1971 serotype C)) | Gaps | Score | Score(bits) |
| 29% | 48% | 615 | 644 | 55 | 646 | 253 |
Local alignment with maximum score contains a part of the 3CD protein (1566:2209) from Poliovirus and Picornain 3C(1646:1858) with RNA-directed RNA polymerase 3D-POL(1859:2328) from Apthovirus. Second score alignment contains whole Protein 2B(1031:1127) and the biggest part of Protein 2C(1128:1456) from Poliovirus and Apthovirus both.
Scores compairment
| ID | Optimal score | Median | Top quartile | Bits score | p-value |
| PURT_ECOLI with QCRA_BACSU (putatively non-homological) | 36.0 | 33,75 | 36,5 | -5,45 | 43,85 |
| RBSA_ECOLI with RBSA_BACSU (putatively homological) | 1129 | 72,5 | 79,75 | 146,72 | 6,78*10-45 |
Parameters changing result
Gap extension penalty increased
| ID | Optimal score | Median | Top quartile | Bits score | p-value |
| PURT_ECOLI with QCRA_BACSU (putatively non-homological) | 29,0 | 28,0 | 31,0 | 1,33 | 0,39 |
| RBSA_ECOLI with RBSA_BACSU (putatively homological) | 1136 | 39 | 42 | 366,67 | 4,19*10-111 |
Here we can see that after extension penalty increase score in putatively homological proteins didn't change. The reason is that RBSA_ECOLI with RBSA_BACSU alignment contains only 2 single gaps, so extension increasing didn't cause. Alignment score in putatively non-homological proteins diminished on 7 points only, but alignment length changes dramatically: from 132AA to 8AA.
EBLOSUM30 matrix
| ID | Optimal score | Median | Top quartile | Bits score | p-value |
| PURT_ECOLI with QCRA_BACSU (putatively non-homological) | 177,5 | 155,75 | 163 | 4 | 0,0625 |
| RBSA_ECOLI with RBSA_BACSU (putatively homological) | 1561 | 348,25 | 359,5 | 108,8 | 1,77*10-33 |
Here we can see, that score for both alignments increase. But alignment for putatively homological proteins didn't change, and for putatively non-homological proteins is change. We know for sure, that there mustn't be a long alignment with a huge score for non-homological proteins as a result, so we can say, that BLOSUM30 matrix for non-homological proteins tend to show a false-positive result.
Formula validation
| ID | Scores | Median | Top quartile | Bits score |
| PURT_ECOLI with QCRA_BACSU (putatively non-homological) | range(-0,4:13,4) | 44,5 | 47 | 1,66 |
| RBSA_ECOLI with RBSA_BACSU (putatively homological) | range(-0,2:9,2) | 91,5 | 96 | 1,51 |
To validate formula, it was decided to count bits scores for each random alignment and count an average bit score than. As we can see, average bits scores seem different from the proposed score. It may be caused by wrong proposed number or task misunderstanding.
BLAST: homologues search
| ID/AC | Organism | Identities(%) | Positives(%) | Alignment length(Campylobacter coli) | Alignment length(Organism1) | Gaps | Score | Score(bits) | Except | Coverage(%) |
| CYSM_CAMJE / P71128 | Campylobacter jejuni subsp. jejuni NCTC 11168 = ATCC 700819 | 86% | 94% | 299AA | 299AA | 0 | 1356 | 526 | 0.0 | 100% |
| CYSK_NEIMB / Q7DDL5 | Neisseria meningitidis MC58 | 55% | 69% | 299AA | 307AA | 10 | 742 | 290 | 9*10-97 | 100% |