MAIN
TERMS
PROJECTS
ABOUT
Alignments
Subject characteristics
| Protein | Number of subject sequences | Number of subject sequences with E-value < 0.001 | max. E-value | Limitation |
| cysK Campilobacter coli | 96 | 83 | 4.4 | by E-value |
Alignments seem to be limited by E-value because the number of subject sequences didn't reach the limit, while last E-value number was 4.4 and the next one E-value number equal to 17 was for TdcB protein alignment.
BLAST with different word size
| Protein | Number of subject sequences | Number of subject sequences with E-value < 0.001 | max. E-value | Limitation |
| cysK Campilobacter coli | 367 | 129 | 10 | by subjects number (default) |
The number of sequences with valid E-number (<0.001) and non-valid number increase both. It must be a result of a more qualitative search.
BLAST with different search region
| Taxones | Not specified | Bacteria (taxid:2) | Terrabacteria group (taxid:1783272) | Actinobacteria (taxid:201174) | Corynebacteriales (taxid:85007) | Mycobacteriaceae (taxid:1762) | Mycobacterium (taxid:1763) | Mycobacterium tuberculosis complex (taxid:77643) |
| E-values | 6*10-96 | 3*10-96 | 9*10-97 | 2*10-97 | 1*10-97 | 8*10-98 | 8*10-98 | 3*10-98 |
According to Karlin's formula, E-values are in linear dependency with bank size. So we can estimate differences in bank's sizes as differences in E-values sizes. That rough estimation is quite correct as we can see in search summary (image below).
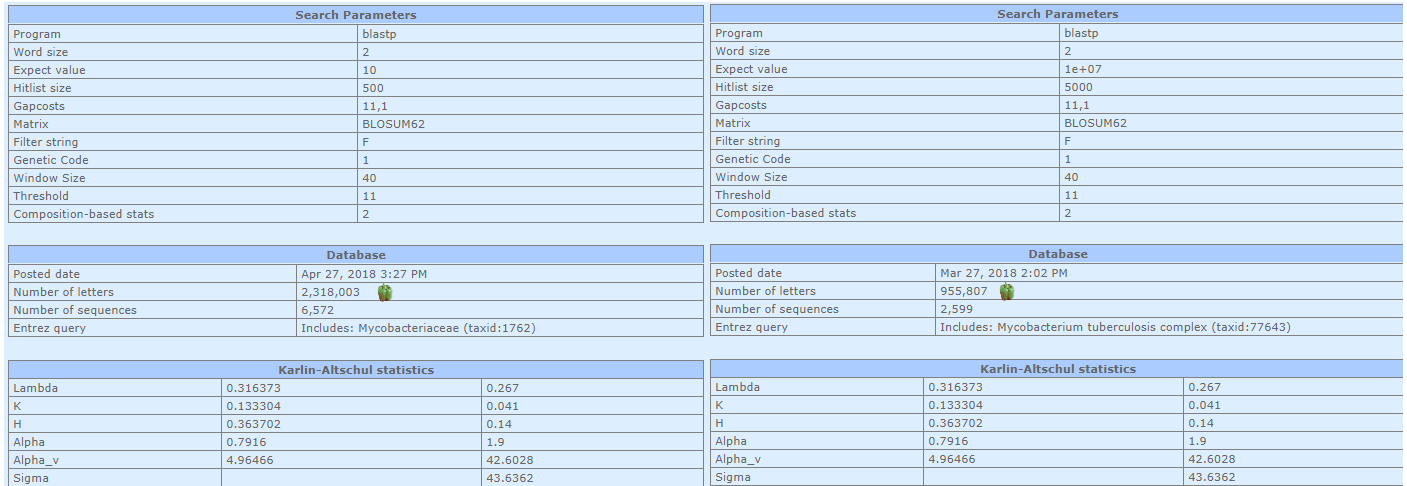
Comparison of BLAST result for different taxonomy. Numbers of letters marked with pepper. On the left side, Mycobacteriaceae family is shown, on the right side is Mycobacterium tuberculosis complex group. The ratio between numbers of letters is 2,42 and between E-values is 2,66.
Calculations with formula show the same result as in search summary. Changing of search capacity won't have an influence on the score, because score number depends on matrix only.
Other BLASTP web-interfaces
| Interfaces/Functions | NCBI | UniProt | EMBL-EBI |
| Input sequence | FASTA/FASTA without definition/sequence portion | FASTA/FASTA without definition/sequence portion | GCG/FASTA/Nucleotide only/GenBank/PIR/NBRF/PHYLIP/Protein only |
| Query subrange | + | - | + |
| Upload file | + | - | + |
| Job title | + | - | - |
| Two and more sequences alignment | + | - | - |
| Databases (for proteins) | Non-redundant sequences, RefSeq, Model organisms, Swiss-prot, Patented sequences, PDB, Metagenomic proteins, Shotgun assembly proteins | UniProtKB, UniRef, UniParc | UniProtKB, UniRef, UniParc, Patented sequences, Other Databases |
| Taxonomy search | Manual taxonomy suggestion | UniProtKB taxonomic subsets only | UniProtKB taxonomic subsets only |
| Entrez query | + | - | - |
| Programs | BLASTP, Quick BLASTP, PSI-BLAST, PHI-BLAST, DELTA-BLAST | BLASTP | BLASTX, BLASTP |
| Parameters | Max target, E-value, Word size, Matrix, Gap costs, compositional adjustments, low complexity regions, masks. | Max target, E-value, Matrix (less than in other tools), low complexity regions, alignment without gaps | Max target, E-value, Word size, Matrix, Gap costs, compositional adjustments, low complexity regions, masks, alignment without gaps, "dropoff" filter, scores filter |
| Results | In current window | In current window/ In new window | In current window/notified by email |
Special aspects of output
NCBI
Karlin-Altschul statistics, MSA viewer, Distance tree of results, Taxonomy reports.
UniProt
Color by % Identity, Taxonomy reports.
EMBL-EBI
Different tools support (Kalign, MAFFT, MView etc.), functional predictions, launched and end date supported.
Recommendations
.NCBI web interface seems pretty useful for some kind of challenges, taxonomy search for example. It also supports entrez queries and different BLAST programs. Otherwise, some minuses are presented such as the inability to work with different formats of queries and current window results opening. EMBL-EBI provides to work with different formats and, in my humble opinion, have better output interface and more useful tools in output. Also, it allows use email notifications. On the other hand, EMBL-EBI interface doesn't support taxonomy suggestion. Both of that interfaces are suited for educational exercises. EMBL-EBI interface is also suited for professional use. UniProt web interface is useless: it doesn't support a lot of tools and isn't suited for professional research.
Obsolete matrix
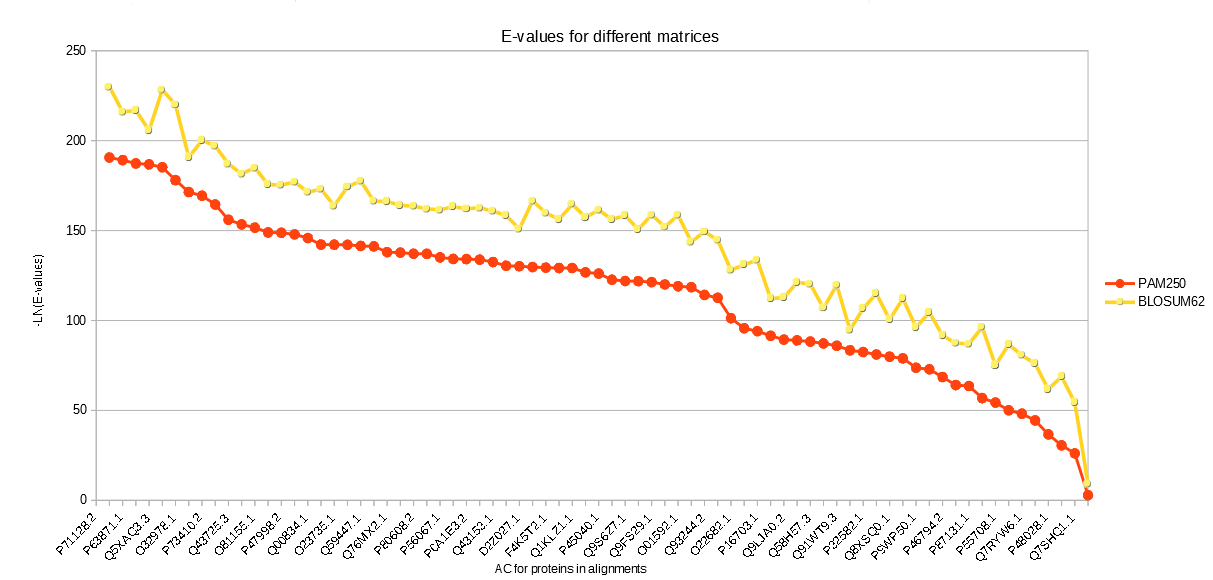
To check the hypothesis that the total number of matches and E-value for good matches will decrease with obsolete matrix search, it was decided to repeat the previous search. It was shown in the output that the total number of alignments dropped from 88 with BLOSUM62 to 78 with PAM250. To check the second part of the hypothesis, it was decided to compare E-values for alignments with the same AC, but different matrices. Results are presented in the graph below: