DNA and RNA structures
Different DNA formes
Different DNA formes
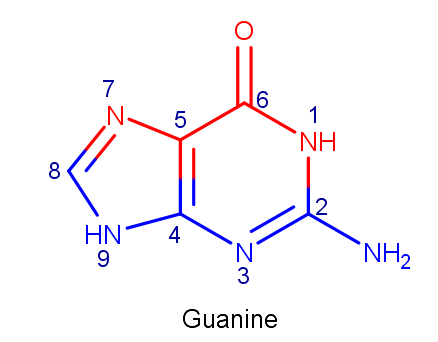
For the second task, B-form of DNA was chosen. Here we can see, that N1, C6, O6, C5, and N7 atoms point out on a major groove, while C2, N2, N3, C4, N9, and C8 atoms point out on a minor groove.
Information table
|
A-form |
B-form |
Z-form |
| Helix type |
Right |
Right |
Left |
| Pitch of a helix |
28.02 |
33.6 |
43.5 |
| BP in a pitch |
11 |
10 |
12 |
| Major groove width |
15.74 (from thymine) |
17.32 (from guanine) |
17.21 (from cytosine) |
| Minor groove width |
11.52 (from cytosine) |
9.99 (from cytosine) |
8.19 (from guanine) |
For the third task, torsion angles in A, B
and Z form of DNA and also tRNA was measured.
Comparing the average angles, we can see, that tRNA is close to A-form.
tRNA angles
Strand I
base alpha beta gamma delta epsilon zeta chi
1 G --- 146.0 56.0 87.3 -171.7 -60.3 -153.6
2 G 141.6 -174.7 -178.4 80.6 -135.9 -65.9 -172.6
3 A -54.1 163.8 53.2 75.6 -166.3 -97.0 -168.1
4 G 156.5 -150.0 -172.9 128.7 -80.3 -46.9 -159.4
5 G 56.0 146.7 51.6 84.1 -134.3 -73.1 177.1
6 C -65.6 165.8 53.8 81.9 -154.7 -73.9 -171.9
7 U -66.3 177.6 55.7 83.3 -157.0 -13.0 -153.8
8 G -137.7 79.1 173.7 88.7 -128.9 -83.0 175.6
9 G 140.9 -141.8 176.1 81.0 -129.1 -79.4 174.0
10 t -62.1 176.5 45.7 78.8 -128.1 -73.1 -162.6
11 P -52.9 163.7 50.7 78.6 -133.3 -90.2 -149.0
12 A -127.0 -103.9 179.0 88.8 -147.2 -70.9 -170.6
13 C -67.2 178.1 47.8 80.0 -160.6 -75.0 -152.9
14 G -66.8 174.4 51.1 82.1 -154.4 -76.4 -156.9
15 A -63.6 166.5 50.4 81.7 -152.6 -72.5 -159.0
16 G -51.3 165.9 52.4 79.1 -162.9 -57.6 -167.9
17 G 160.4 -172.1 179.8 83.8 -142.2 -79.0 179.1
18 G -63.6 170.8 46.7 78.5 -149.7 -76.7 -163.0
19 G 136.0 -128.9 71.0 91.6 -153.1 -68.4 -171.9
20 C -70.2 -179.4 46.6 82.0 -155.0 -69.3 -160.6
21 G -59.0 178.1 43.9 78.6 -159.5 -73.4 -154.6
22 C 148.9 -155.6 -174.5 85.0 -141.5 -77.5 -174.1
23 A -71.4 167.8 64.3 75.7 -152.6 -61.5 -174.0
24 G -59.7 177.2 54.1 87.7 -128.7 -87.8 -150.9
25 G -63.4 -158.5 46.5 126.7 -173.7 145.9 -71.5
Strand II
base alpha beta gamma delta epsilon zeta chi
1 C -63.8 170.5 54.8 81.4 --- --- -154.8
2 C -68.2 179.3 46.3 80.1 -151.7 -73.5 -156.1
3 U -64.6 -170.0 50.0 84.8 -166.2 -73.7 -158.3
4 C -62.1 177.2 51.6 80.3 -157.6 -61.1 -171.4
5 U -74.0 171.3 59.2 82.3 -152.7 -79.9 -168.3
6 G -62.5 160.5 63.5 79.2 -156.2 -76.5 -168.6
7 A -64.5 177.5 52.1 80.4 -146.1 -72.5 -163.5
8 C -65.5 -178.8 49.9 83.3 -151.4 -72.9 -164.4
9 C -122.5 -136.8 51.9 81.7 -156.2 -73.6 -166.8
10 G -48.4 -146.4 56.5 139.2 -107.8 -150.2 -66.7
11 G -51.6 146.1 145.4 132.3 -137.3 -68.4 -135.1
12 U -71.7 171.9 61.3 79.5 -162.7 -39.6 -157.2
13 G -72.7 164.2 63.8 80.7 -159.6 -67.8 -161.1
14 C -70.5 177.3 51.7 80.2 -153.3 -71.7 -152.7
15 U -64.3 -174.8 48.9 80.8 -152.4 -69.8 -170.1
16 C -70.5 -168.8 47.7 82.9 -154.1 -68.8 -166.4
17 C -61.4 176.6 46.2 81.7 -154.1 -73.2 -161.0
18 A -59.9 173.8 49.7 84.8 -142.6 -68.9 -162.9
19 C -67.1 171.6 52.1 80.9 -145.3 -66.9 -162.2
20 G -70.5 177.8 51.6 82.5 -142.3 -72.1 -166.6
21 C -75.4 178.3 53.8 79.2 -158.5 -76.3 -160.1
22 G -53.8 159.3 57.3 82.1 -160.8 -75.6 -164.8
23 U -91.1 -144.8 65.8 83.9 -141.9 -133.0 -167.8
24 C -53.5 134.3 51.0 129.4 -131.9 73.1 -138.5
25 C 157.0 -167.8 59.7 85.3 -141.6 -62.3 -162.6
tRNA stems
Strand I Strand II Helix
1 (0.008) ....>T:...4_:[..G]G-----C[..C]:..69_:T<.... (0.008) |
2 (0.009) ....>T:...5_:[..G]G-----C[..C]:..68_:T<.... (0.006) |
3 (0.012) ....>T:...6_:[..A]A-----U[..U]:..67_:T<.... (0.003) |
4 (0.005) ....>T:...7_:[..G]G-----C[..C]:..66_:T<.... (0.010) |
5 (0.005) ....>T:..49_:[..G]G-*---U[..U]:..65_:T<.... (0.010) |
6 (0.004) ....>T:..50_:[..C]C-----G[..G]:..64_:T<.... (0.013) |
7 (0.002) ....>T:..51_:[..U]U-----A[..A]:..63_:T<.... (0.004) |
8 (0.006) ....>T:..52_:[..G]G-----C[..C]:..62_:T<.... (0.002) |
9 (0.005) ....>T:..53_:[..G]G-----C[..C]:..61_:T<.... (0.005) |
12 (0.011) ....>T:..38_:[..A]A-----U[..U]:..32_:T<.... (0.009) |
13 (0.003) ....>T:..39_:[..C]C-----G[..G]:..31_:T<.... (0.006) |
14 (0.006) ....>T:..40_:[..G]G-----C[..C]:..30_:T<.... (0.007) |
15 (0.008) ....>T:..41_:[..A]A-----U[..U]:..29_:T<.... (0.009) |
16 (0.005) ....>T:..42_:[..G]G-----C[..C]:..28_:T<.... (0.005) |
17 (0.008) ....>T:..43_:[..G]G-----C[..C]:..27_:T<.... (0.007) |
18 (0.007) ....>T:..44_:[..G]G-**--A[..A]:..26_:T<.... (0.008) |
19 (0.004) ....>T:..10_:[..G]G-----C[..C]:..26_:T<.... (0.006) |
20 (0.003) ....>T:..11_:[..C]C-----G[..G]:..25_:T<.... (0.004) |
21 (0.004) ....>T:..12_:[..G]G-----C[..C]:..24_:T<.... (0.003) |
22 (0.004) ....>T:..13_:[..C]C-----G[..G]:..23_:T<.... (0.005) |
non-Watson-Crick base-pairs
5 (0.005) ....>T:..49_:[..G]G-*---U[..U]:..65_:T<.... (0.010) |
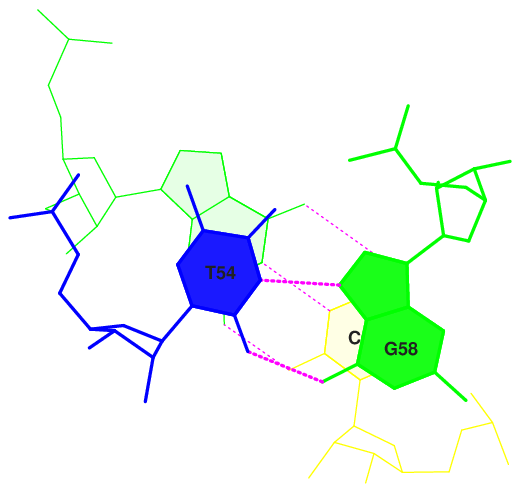
10 (0.006) ....>T:..54_:[5MU]t-**--G[..G]:..58_:T<.... (0.006) |
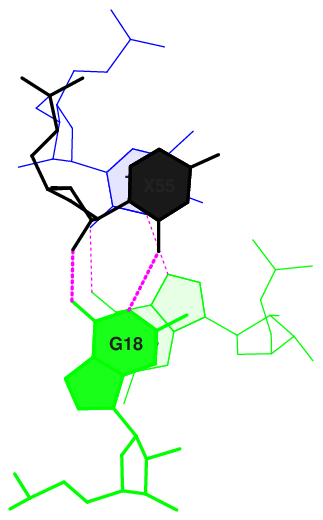
11 (0.026) ....>T:..55_:[PSU]P-**+-G[..G]:..18_:T<.... (0.012) x
18 (0.007) ....>T:..44_:[..G]G-**--A[..A]:..26_:T<.... (0.008) |
23 (0.003) ....>T:..14_:[..A]A-**--U[..U]:...8_:T<.... (0.004) |
24 (0.004) ....>T:..15_:[..G]G-**+-C[..C]:..48_:T<.... (0.007) x
additional H-bonds
25 (0.011) ....>T:..19_:[..G]G-----C[..C]:..56_:T<.... (0.005) +
Stacking interactions
Here overlap area in Å 2 is presented. We can see, that the highest overlap area is shown for 9 and 10 (Gt/GC and tP/GG) pairs.
step i1-i2 i1-j2 j1-i2 j1-j2 sum
1 GG/CC 3.88( 2.64) 0.00( 0.00) 0.05( 0.00) 0.57( 0.02) 4.50( 2.66)
2 GA/UC 5.04( 3.03) 0.00( 0.00) 0.00( 0.00) 0.08( 0.00) 5.12( 3.03)
3 AG/CU 1.42( 1.41) 0.00( 0.00) 1.21( 0.00) 0.00( 0.00) 2.63( 1.41)
4 GG/UC 1.59( 0.26) 0.00( 0.00) 1.10( 0.00) 0.00( 0.00) 2.69( 0.26)
5 GC/GU 6.56( 3.49) 0.00( 0.00) 0.00( 0.00) 6.55( 3.64) 13.11( 7.12)
6 CU/AG 0.00( 0.00) 0.00( 0.00) 0.38( 0.00) 3.82( 3.26) 4.20( 3.26)
7 UG/CA 0.00( 0.00) 0.00( 0.00) 4.80( 3.05) 0.00( 0.00) 4.80( 3.05)
8 GG/CC 5.09( 2.72) 0.00( 0.00) 0.74( 0.00) 0.00( 0.00) 5.83( 2.72)
9 Gt/GC 7.60( 2.39) 0.00( 0.00) 0.00( 0.00) 5.65( 2.63) 13.24( 5.01)
10 tP/GG 7.22( 2.93) 0.00( 0.00) 0.00( 0.00) 6.02( 3.11) 13.24( 6.04)
11 PA/UG 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
12 AC/GU 3.07( 1.61) 0.00( 0.00) 0.00( 0.00) 7.18( 4.20) 10.25( 5.81)
13 CG/CG 0.52( 0.00) 0.00( 0.00) 2.89( 0.58) 1.19( 0.19) 4.59( 0.77)
14 GA/UC 4.54( 3.06) 0.00( 0.00) 0.00( 0.00) 1.33( 0.11) 5.87( 3.17)
15 AG/CU 2.91( 2.55) 0.00( 0.00) 0.58( 0.00) 0.00( 0.00) 3.50( 2.55)
16 GG/CC 3.57( 1.78) 0.00( 0.00) 1.11( 0.00) 0.00( 0.00) 4.68( 1.78)
17 GG/AC 1.05( 0.00) 0.00( 0.00) 0.00( 0.00) 2.78( 1.53) 3.82( 1.53)
18 GG/CA 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
19 GC/GC 4.70( 1.85) 0.00( 0.00) 0.00( 0.00) 7.21( 4.46) 11.91( 6.31)
20 CG/CG 0.38( 0.00) 0.00( 0.00) 4.44( 1.72) 0.00( 0.00) 4.83( 1.72)
21 GC/GC 6.58( 2.45) 0.00( 0.00) 0.00( 0.00) 3.40( 0.98) 9.98( 3.44)
22 CA/UG 0.00( 0.00) 1.35( 0.00) 4.78( 1.78) 0.00( 0.00) 6.13( 1.78)
23 AG/CU 3.84( 1.22) 0.00( 0.00) 0.02( 0.00) 0.00( 0.00) 3.86( 1.22)
24 GG/CC 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
The strongest overlaps are visualised below.
© Gumerov Ruslan, 2017