Sanger sequencing
Sanger sequences analysis
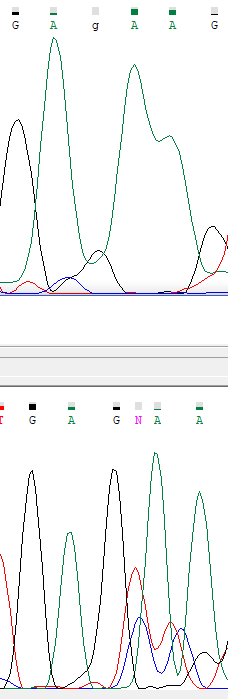
In forward sequence we can see that between 2 adenines there is one uninterpretable peak, corresponding to be a guanine. In reverse sequence between 2 adenines, there are 2 peaks, one of them is definitely
a guanine spectrum, and another one is an uninterpretable peak. It seems that the original sequence has a "GAGAAG" pattern in that place, but in forward read guanine peak missed, and in reverse read,
new peaks incorporated.
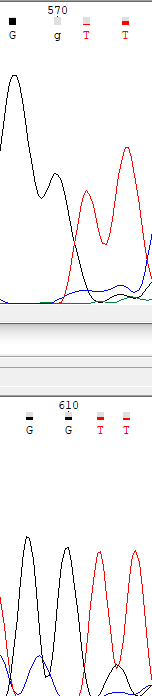
Here we can see, that in forward sequence analyzer didn't recognize second guanine peak, while in reverse sequence both peaks recognized.
In that part of a sequence, cytosine peak in forward read didn't recognize because of low signal-to-noise ratio.
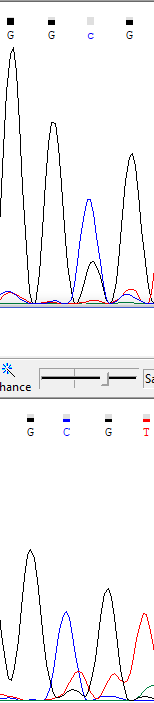
Here we can see, that analyzer didn't recognize an adenine in low complexity region, containing adenines.
In the forward sequence, without any editing, the coordinates of uninterpretable regions are 0;27 and 650; 691. In reverse sequence, coordinates, according
to the forward sequence, are -40;-19 and 674;696. Signal-to-noise ratio for the current sequence is quite low, I consider it appropriate for future studies. The highest amount of noise localized at the
beginning and at the end of a sequence, and that is ordinary for that sequence method.
"Bad" sequences
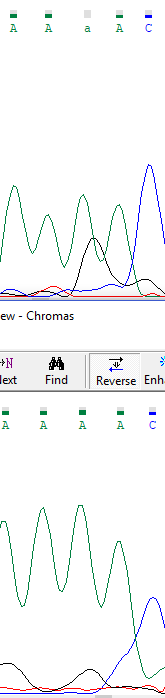
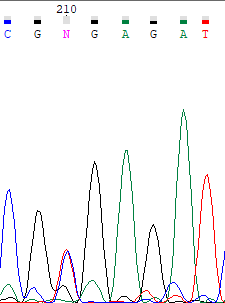
In that sequence, it must be a polymorphism in 210 position (2 similar peaks of timine and cytosine in the same position are shown, without any noise surround).
© Gumerov Ruslan, 2017