MAIN
TERMS
PROJECTS
ABOUT
Cellulase/esterase CelE
Cellulase/esterase CelE is a multifunctional enzyme involved in the degradation of plant cell wall polysaccharides.
It catalyzes Endohydrolysis of (1->4)-beta-D-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans,
and also Deacetylation of xylans and xylo-oligosaccharides, and involves, in general, in cellulose degradation pathway
and xylan degradation pathway both. Cellulase/esterase CelE is a sectetory protein located in the extracellular region.
UniProt AC p10477 is a Cellulase/esterase CelE from the bacterium Clostridium thermocellum strain ATCC 27405.
Information table
| UniProt ID | CELE_CLOTH |
| UniProt AC | P10477 |
| RefSeq ID | WP_011837903.1 |
| PDB ID | 2WAB;2WAO;4IM4 |
| Length | 814 AA |
| Weight | 90230 Da |
| RecName | Full=Cellulase/esterase CelE |
Secondary structure isn't shown for the whole protein. PDB contains 3 entries of a Cellulase/esterase CelE, both of them are identified with X-ray metod. The newest 4IM4 entry include 6 chains, while the others displays only one.
UniRef Clusters
UniRef100_P10477 cluster contains only Cellulase/esterase CelE protein. UniRef90_P10477 cluster, as well as the UniRef50_P10477 cluster, contains 4 cluster members: p10477, UPI00017E2E24, UPI0001B1EF96 and UPI0003B8D537, while only p10477 got a Swiss-Prot reviewed status with the maximum score.
Exploring the UniProt
-
name:"cellulase esterase cele"
In the output 5 proteins are shown, while Cellulase/esterase CelE with AC p10477 only reviewed by Swiss-Prot, others are predicted or inferred from homology. 3 from other entries correspond to different organisms from Fungi clade, and the last AC is from an unclassified bacterium HR30. Both of proteins shows an ester hydrolase activity.
-
organism:"clostridium thermocellum" name:"cellulase esterase cele"
3 different strains of Clostridium Thermocellum are presented in the UniProt, but only one entry is shown in the output, which refers us that only one cellulase esterase exists in Clostridium Thermocellum strain ATCC 27405 proteome. One from the other strains has only 3 unreviewed entries, while the last one is quiet big, but do not include Cellulase/esterase either.
-
taxonomy:ruminococcaceae name:"cellulase esterase cele"
Only 1 entry with AC p10477 is presented in the output.
-
taxonomy:firmicutes name:"cellulase esterase cele"
Only 1 entry with AC p10477 is presented in the output.
-
name:homeobox
44,012 entries are presented in the output, 1396 from them are reviewed. Homeobox proteins present in different organisms, Mus musculus,Homo sapiens, Arabidopsis thaliana etc. They are involved in the regulation of anatomical development.
-
name:homeobox taxonomy:ciliophora
2 unreviewed entries are presented in the output. One of them is a "Homeobox protein bel1-like protein" from a Stylonychia lemnae, and the other is "Homeobox protein, putative" from Tetrahymena thermophila. Both of them are predicted proteins.
-
name:homeobox taxonomy:Fungi
1,339 entries are presented in the output, 18 from them are reviewed.
-
name:trypsin
18,951 entries are presented in the output, 311 from them are reviewed.
-
name:trypsin name:inhibitor
3,914 entries are presented in the output, 210 from them are reviewed.
Comparing RefSeq with UniProt
UniProt and RefSeq entries look quite similar, but some differences should be matched either. UniProt entry contains more information about protein on single page. For instance, kinetics, pathways and cellular location are shown, which makes UniProt entry easier to understand. It seems that reviewed UniProt entry provide by an inspector, so much more data from publications accessible on the entry's page. Also, validating data in UniProt is happening more often, several times a year, while on the RefSeq database last update of analyzed protein was in 2013. Recommended by UniProt protein name and taxonomical name of a clade are differ from the same parameters in RefSeq: type of a protein (glycoside hydrolase) is pointed out as a protein name. But, surprisingly, RefSeq searching clusters tool shows a cluster with 5 proteins conserved in Ruminococcaceae family with that type name, which makes RefSeq database more useful in this case. Also, it's easily to notice the lack of a links to the different resources in RefSeq: UniProt shows ID's from different databases (eggNOG, CDD, Pfam) on the one single page.
History of an entry
The most noticeable update was, of course, update of a hypertext transfer protocol.
 Comparing with a first version of
an entry, we can see a vast increase of a publications about protein. Also, second sequence was made in 2015, while AC A3DDK3 was replaced and first
sequence mistake (replascing valine on leucine)was shown.
Comparing with a first version of
an entry, we can see a vast increase of a publications about protein. Also, second sequence was made in 2015, while AC A3DDK3 was replaced and first
sequence mistake (replascing valine on leucine)was shown.  Includes, different functions, secondary structure and kinetic parameters was defined either.
Includes, different functions, secondary structure and kinetic parameters was defined either.


Unstandard modifications
All of the unstandard modifications describe in the "FT" strings (in text format). On html page each of the occurrences got it's own container (see images below).
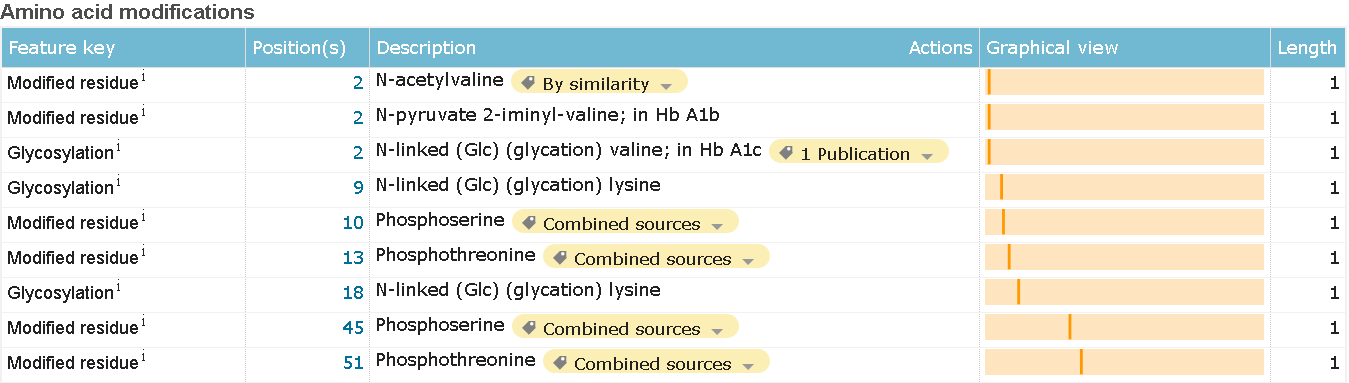 Modified residue and glycosylation
Modified residue and glycosylation
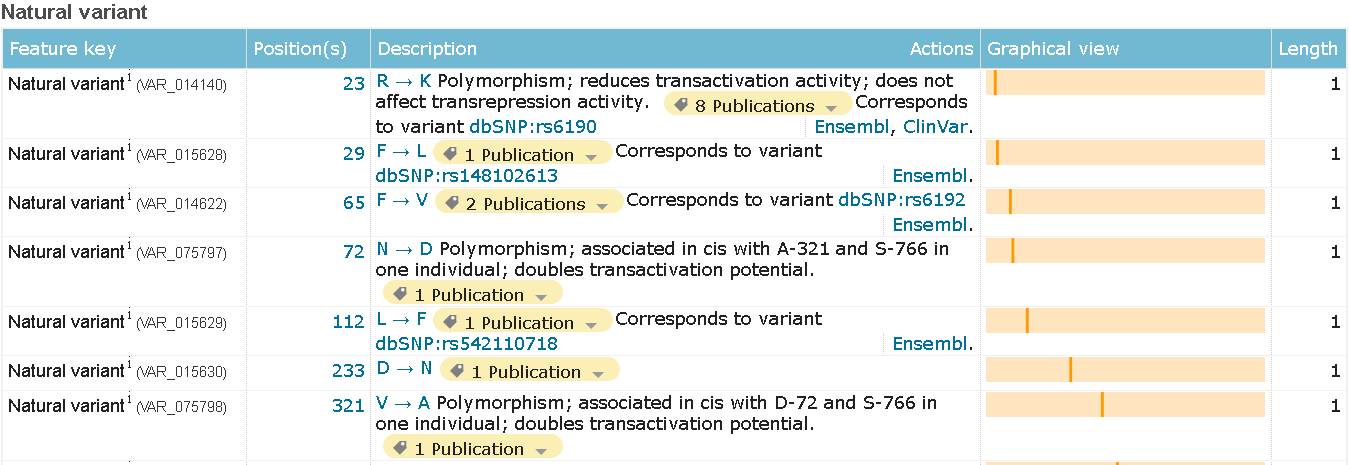 Polymorphism
Polymorphism
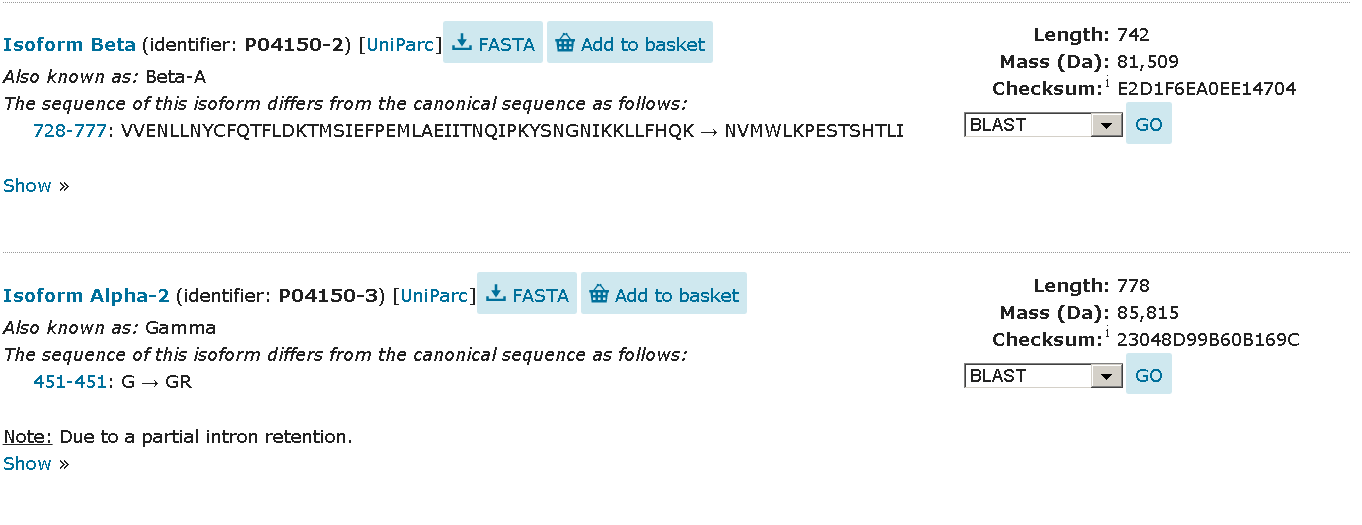 Alternative splicing isoforms
Alternative splicing isoforms
 Non-standard residues
Non-standard residues
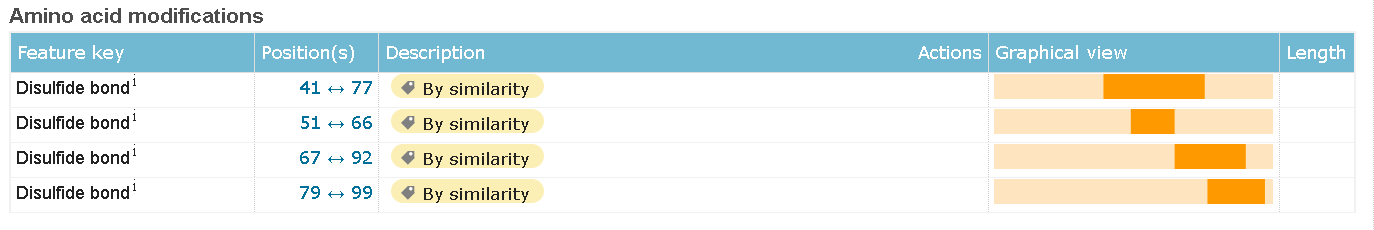 Disulfide bonds
Disulfide bonds
 Modified residue and glycosylation
Modified residue and glycosylation Polymorphism
Polymorphism Alternative splicing isoforms
Alternative splicing isoforms Non-standard residues
Non-standard residues  Disulfide bonds
Disulfide bonds