*****************
* O R C A *
*****************
--- An Ab Initio, DFT and Semiempirical electronic structure package ---
#######################################################
# -***- #
# Department of molecular theory and spectroscopy #
# Directorship: Frank Neese #
# Max Planck Institute for Chemical Energy Conversion #
# D-45470 Muelheim/Ruhr #
# Germany #
# #
# All rights reserved #
# -***- #
#######################################################
Program Version 3.0.3 - RELEASE -
With contributions from (in alphabetic order):
Ute Becker : Parallelization
Dmytro Bykov : SCF Hessian
Dmitry Ganyushin : Spin-Orbit,Spin-Spin,Magnetic field MRCI
Andreas Hansen : Spin unrestricted coupled pair/coupled cluster methods
Dimitrios Liakos : Extrapolation schemes; parallel MDCI
Robert Izsak : Overlap fitted RIJCOSX, COSX-SCS-MP3
Christian Kollmar : KDIIS, OOCD, Brueckner-CCSD(T), CCSD density
Simone Kossmann : Meta GGA functionals, TD-DFT gradient, OOMP2, MP2 Hessian
Taras Petrenko : DFT Hessian,TD-DFT gradient, ASA and ECA modules, normal mode analysis, Resonance Raman, ABS, FL, XAS/XES, NRVS
Christoph Reimann : Effective Core Potentials
Michael Roemelt : Restricted open shell CIS
Christoph Riplinger : Improved optimizer, TS searches, QM/MM, DLPNO-CCSD
Barbara Sandhoefer : DKH picture change effects
Igor Schapiro : Molecular dynamics
Kantharuban Sivalingam : CASSCF convergence, NEVPT2
Boris Wezisla : Elementary symmetry handling
Frank Wennmohs : Technical directorship
We gratefully acknowledge several colleagues who have allowed us to
interface, adapt or use parts of their codes:
Stefan Grimme, W. Hujo, H. Kruse, T. Risthaus : VdW corrections, initial TS optimization,
DFT functionals, gCP
Ed Valeev : LibInt (2-el integral package), F12 methods
Garnet Chan, S. Sharma, R. Olivares : DMRG
Ulf Ekstrom : XCFun DFT Library
Mihaly Kallay : mrcc (arbitrary order and MRCC methods)
Andreas Klamt, Michael Diedenhofen : otool_cosmo (COSMO solvation model)
Frank Weinhold : gennbo (NPA and NBO analysis)
Christopher J. Cramer and Donald G. Truhlar : smd solvation model
Your calculation uses the libint2 library for the computation of 2-el integrals
For citations please refer to: http://libint.valeyev.net
This ORCA versions uses:
CBLAS interface : Fast vector & matrix operations
LAPACKE interface : Fast linear algebra routines
SCALAPACK package : Parallel linear algebra routines
Your calculation utilizes the basis: Ahlrichs-VDZ
Cite in your paper:
H - Kr: A. Schaefer, H. Horn and R. Ahlrichs, J. Chem. Phys. 97, 2571 (1992).
Your calculation utilizes the basis: Ahlrichs SVPalls1+f
Cite in your paper:
Rb - Xe: A. Schaefer, C. Huber and R. Ahlrichs, J. Chem. Phys. 100, 5829 (1994).
Your calculation utilizes pol. fcns from basis: Ahlrichs polarization
Cite in your paper:
H - Kr: R. Ahlrichs and coworkers, unpublished
================================================================================
WARNINGS
Please study these warnings very carefully!
================================================================================
Now building the actual basis set
INFO : the flag for use of LIBINT has been found!
================================================================================
INPUT FILE
================================================================================
NAME = h.inp
| 1> ! UHF SVP XYZFile
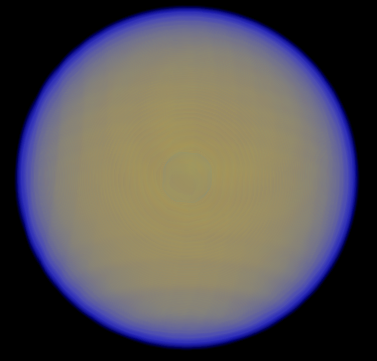
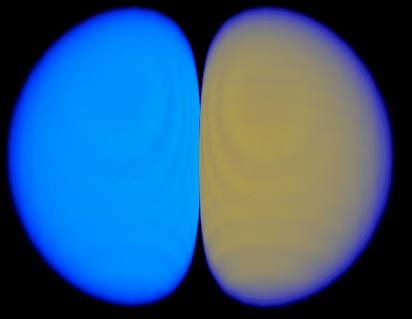
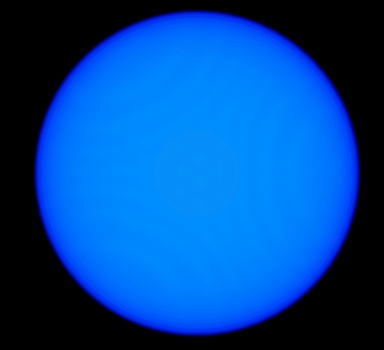
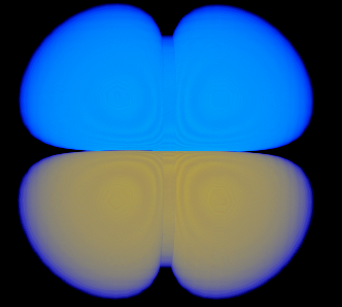
| 2> %plots Format Cube
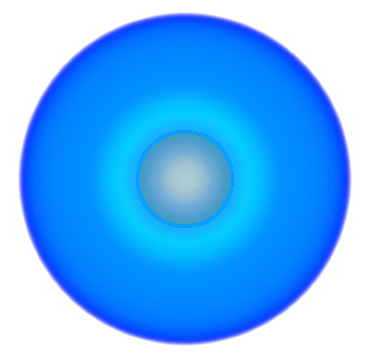
| 3> MO("H-1.cube",1,0);
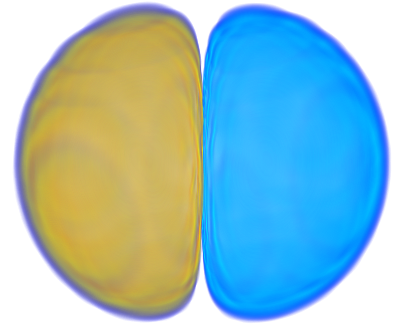
| 4> MO("H-2.cube",2,0);
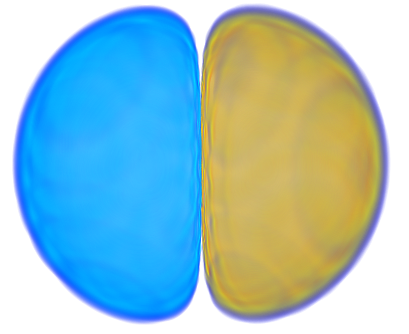
| 5> MO("H-3.cube",3,0);
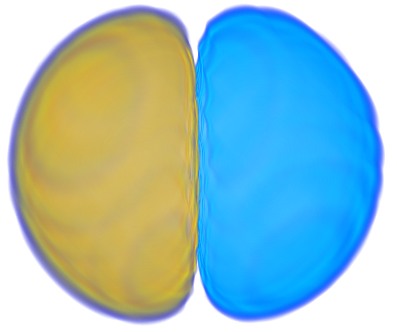
| 6> MO("H-4.cube",4,0);
| 7> end
| 8> * xyz 0 4
| 9> H 0 0 0
| 10> ** ****END OF INPUT****
================================================================================
****************************
* Single Point Calculation *
****************************
---------------------------------
CARTESIAN COORDINATES (ANGSTROEM)
---------------------------------
H 0.000000 0.000000 0.000000
----------------------------
CARTESIAN COORDINATES (A.U.)
----------------------------
NO LB ZA FRAG MASS X Y Z
0 H 1.0000 0 1.008 0.000000000000000 0.000000000000000 0.000000000000000
--------------------------------
INTERNAL COORDINATES (ANGSTROEM)
--------------------------------
H 0 0 0 0.000000 0.000 0.000
---------------------------
INTERNAL COORDINATES (A.U.)
---------------------------
H 0 0 0 0.000000 0.000 0.000
---------------------
BASIS SET INFORMATION
---------------------
There are 1 groups of distinct atoms
Group 1 Type H : 4s1p contracted to 2s1p pattern {31/1}
Atom 0H basis set group => 1
------------------------------------------------------------------------------
ORCA GTO INTEGRAL CALCULATION
------------------------------------------------------------------------------
BASIS SET STATISTICS AND STARTUP INFO
# of primitive gaussian shells ... 5
# of primitive gaussian functions ... 7
# of contracted shell ... 3
# of contracted basis functions ... 5
Highest angular momentum ... 1
Maximum contraction depth ... 3
Integral package used ... LIBINT
Integral threshhold Thresh ... 1.000e-10
Primitive cut-off TCut ... 1.000e-11
INTEGRAL EVALUATION
One electron integrals ... done
Pre-screening matrix ... done
Shell pair data ... done ( 0.000 sec)
-------------------------------------------------------------------------------
ORCA SCF
-------------------------------------------------------------------------------
------------
SCF SETTINGS
------------
Hamiltonian:
Ab initio Hamiltonian Method .... Hartree-Fock(GTOs)
General Settings:
Integral files IntName .... h
Hartree-Fock type HFTyp .... UHF
Total Charge Charge .... 0
Multiplicity Mult .... 4
Number of Electrons NEL .... 1
Basis Dimension Dim .... 5
Nuclear Repulsion ENuc .... 0.0000000000 Eh
Convergence Acceleration:
DIIS CNVDIIS .... on
Start iteration DIISMaxIt .... 12
Startup error DIISStart .... 0.200000
# of expansion vecs DIISMaxEq .... 5
Bias factor DIISBfac .... 1.050
Max. coefficient DIISMaxC .... 10.000
Newton-Raphson CNVNR .... off
SOSCF CNVSOSCF .... off
Level Shifting CNVShift .... on
Level shift para. LevelShift .... 0.2500
Turn off err/grad. ShiftErr .... 0.0010
Zerner damping CNVZerner .... off
Static damping CNVDamp .... on
Fraction old density DampFac .... 0.7000
Max. Damping (<1) DampMax .... 0.9800
Min. Damping (>=0) DampMin .... 0.0000
Turn off err/grad. DampErr .... 0.1000
Fernandez-Rico CNVRico .... off
SCF Procedure:
Maximum # iterations MaxIter .... 125
SCF integral mode SCFMode .... Direct
Integral package .... LIBINT
Reset frequeny DirectResetFreq .... 20
Integral Threshold Thresh .... 1.000e-10 Eh
Primitive CutOff TCut .... 1.000e-11 Eh
Convergence Tolerance:
Convergence Check Mode ConvCheckMode .... Total+1el-Energy
Energy Change TolE .... 1.000e-06 Eh
1-El. energy change .... 1.000e-03 Eh
DIIS Error TolErr .... 1.000e-06
Diagonalization of the overlap matrix:
Smallest eigenvalue ... 3.152e-01
Time for diagonalization ... 0.000 sec
Threshold for overlap eigenvalues ... 1.000e-08
Number of eigenvalues below threshold ... 0
Time for construction of square roots ... 0.000 sec
Total time needed ... 0.001 sec
-------------------
DFT GRID GENERATION
-------------------
General Integration Accuracy IntAcc ... 4.340
Radial Grid Type RadialGrid ... Gauss-Chebyshev
Angular Grid (max. acc.) AngularGrid ... Lebedev-110
Angular grid pruning method GridPruning ... 3 (G Style)
Weight generation scheme WeightScheme... Becke
Basis function cutoff BFCut ... 1.0000e-10
Integration weight cutoff WCut ... 1.0000e-14
Grids for H and He will be reduced by one unit
# of grid points (after initial pruning) ... 794 ( 0.0 sec)
# of grid points (after weights+screening) ... 794 ( 0.0 sec)
Grid point division into batches done ... 0.0 sec
Reduced shell lists constructed in 0.0 sec
Total number of grid points ... 794
Total number of batches ... 13
Average number of points per batch ... 61
Average number of grid points per atom ... 794
Average number of shells per batch ... 2.79 (92.86%)
Average number of basis functions per batch ... 4.64 (92.86%)
Average number of large shells per batch ... 2.79 (100.00%)
Average number of large basis fcns per batch ... 4.64 (100.00%)
Maximum spatial batch extension ... 17.62, 21.59, 21.59 au
Average spatial batch extension ... 6.10, 8.93, 9.49 au
Time for grid setup = 0.003 sec
------------------------------
INITIAL GUESS: MODEL POTENTIAL
------------------------------
Loading Hartree-Fock densities ... done
Calculating cut-offs ... done
Setting up the integral package ... done
Initializing the effective Hamiltonian ... done
Starting the Coulomb interaction ... done ( 0.0 sec)
Reading the grid ... done
Mapping shells ... done
Starting the XC term evaluation ... done ( 0.0 sec)
Transforming the Hamiltonian ... done ( 0.0 sec)
Diagonalizing the Hamiltonian ... done ( 0.0 sec)
Back transforming the eigenvectors ... done ( 0.0 sec)
Now organizing SCF variables ... done
------------------
INITIAL GUESS DONE ( 0.2 sec)
------------------
--------------
SCF ITERATIONS
--------------
ITER Energy Delta-E Max-DP RMS-DP [F,P] Damp
*** Starting incremental Fock matrix formation ***
0 0.0728625057 0.000000000000 0.00000000 0.00000000 0.0000000 0.7000
**** Energy Check signals convergence ****
*****************************************************
* SUCCESS *
* SCF CONVERGED AFTER 1 CYCLES *
*****************************************************
----------------
TOTAL SCF ENERGY
----------------
Total Energy : 0.07286251 Eh 1.98269 eV
Components:
Nuclear Repulsion : 0.00000000 Eh 0.00000 eV
Electronic Energy : 0.07286251 Eh 1.98269 eV
One Electron Energy: -0.31757803 Eh -8.64174 eV
Two Electron Energy: 0.39044053 Eh 10.62443 eV
Virial components:
Potential Energy : -1.57795800 Eh -42.93842 eV
Kinetic Energy : 1.65082051 Eh 44.92111 eV
Virial Ratio : 0.95586285
---------------
SCF CONVERGENCE
---------------
Last Energy change ... -5.5511e-17 Tolerance : 1.0000e-06
Last MAX-Density change ... 2.2204e-16 Tolerance : 1.0000e-05
Last RMS-Density change ... 4.7969e-17 Tolerance : 1.0000e-06
Last DIIS Error ... 1.9938e-16 Tolerance : 1.0000e-06
**** THE GBW FILE WAS UPDATED (h.gbw) ****
**** DENSITY FILE WAS UPDATED (h.scfp.tmp) ****
**** ENERGY FILE WAS UPDATED (h.en.tmp) ****
----------------------
UHF SPIN CONTAMINATION
----------------------
Expectation value of <S**2> : 2.000000
Ideal value S*(S+1) for S=1.0 : 2.000000
Deviation : 0.000000
----------------
ORBITAL ENERGIES
----------------
SPIN UP ORBITALS
NO OCC E(Eh) E(eV)
0 1.0000 -0.108838 -2.9616
1 1.0000 0.572141 15.5687
2 0.0000 2.100372 57.1540
3 0.0000 2.100372 57.1540
4 0.0000 2.100372 57.1540
SPIN DOWN ORBITALS
NO OCC E(Eh) E(eV)
0 0.0000 0.487405 13.2630
1 0.0000 1.318415 35.8759
2 0.0000 2.270122 61.7732
3 0.0000 2.270122 61.7732
4 0.0000 2.270122 61.7732
********************************
* MULLIKEN POPULATION ANALYSIS *
********************************
**** WARNING: MULLIKEN FINDS 2.0000000 ELECTRONS INSTEAD OF 1 ****
--------------------------------------------
MULLIKEN ATOMIC CHARGES AND SPIN POPULATIONS
--------------------------------------------
0 H : -1.000000 2.000000
Sum of atomic charges : -1.0000000
Sum of atomic spin populations: 2.0000000
-----------------------------------------------------
MULLIKEN REDUCED ORBITAL CHARGES AND SPIN POPULATIONS
-----------------------------------------------------
CHARGE
0 H s : 2.000000 s : 2.000000
pz : 0.000000 p : 0.000000
px : 0.000000
py : 0.000000
SPIN
0 H s : 2.000000 s : 2.000000
pz : 0.000000 p : 0.000000
px : 0.000000
py : 0.000000
*******************************
* LOEWDIN POPULATION ANALYSIS *
*******************************
**** WARNING: LOEWDIN FINDS 2.0000000 ELECTRONS INSTEAD OF 1 ****
-------------------------------------------
LOEWDIN ATOMIC CHARGES AND SPIN POPULATIONS
-------------------------------------------
0 H : -1.000000 2.000000
----------------------------------------------------
LOEWDIN REDUCED ORBITAL CHARGES AND SPIN POPULATIONS
----------------------------------------------------
CHARGE
0 H s : 2.000000 s : 2.000000
pz : 0.000000 p : 0.000000
px : 0.000000
py : 0.000000
SPIN
0 H s : 2.000000 s : 2.000000
pz : 0.000000 p : 0.000000
px : 0.000000
py : 0.000000
*****************************
* MAYER POPULATION ANALYSIS *
*****************************
NA - Mulliken gross atomic population
ZA - Total nuclear charge
QA - Mulliken gross atomic charge
VA - Mayer's total valence
BVA - Mayer's bonded valence
FA - Mayer's free valence
ATOM NA ZA QA VA BVA FA
0 H 2.0000 1.0000 -1.0000 2.0000 0.0000 2.0000
Mayer bond orders larger than 0.1
-----------------------------------------------
ATOM BASIS FOR ELEMENT H
-----------------------------------------------
NSH[1] = 3;
res=GAUSS_InitGTOSTO(BG,BS,1,NSH[1]);
// Basis function for L=s
(*BG)[ 1][ 0].l = ((*BS)[ 1][ 0].l=0);
(*BG)[ 1][ 0].ng = 4;
(*BG)[ 1][ 0].a[ 0]= 13.01070100; (*BG)[ 1][ 0].d[ 0]= 0.096096678877;
(*BG)[ 1][ 0].a[ 1]= 1.96225720; (*BG)[ 1][ 0].d[ 1]= 0.163022191701;
(*BG)[ 1][ 0].a[ 2]= 0.44453796; (*BG)[ 1][ 0].d[ 2]= 0.185592186247;
(*BG)[ 1][ 0].a[ 3]= 0.12194962; (*BG)[ 1][ 0].d[ 3]= 0.073701452542;
// Basis function for L=s
(*BG)[ 1][ 1].l = ((*BS)[ 1][ 1].l=0);
(*BG)[ 1][ 1].ng = 4;
(*BG)[ 1][ 1].a[ 0]= 13.01070100; (*BG)[ 1][ 1].d[ 0]= 0.202726593388;
(*BG)[ 1][ 1].a[ 1]= 1.96225720; (*BG)[ 1][ 1].d[ 1]= 0.343913379281;
(*BG)[ 1][ 1].a[ 2]= 0.44453796; (*BG)[ 1][ 1].d[ 2]= 0.391527283951;
(*BG)[ 1][ 1].a[ 3]= 0.12194962; (*BG)[ 1][ 1].d[ 3]=-0.187888280974;
// Basis function for L=p
(*BG)[ 1][ 2].l = ((*BS)[ 1][ 2].l=1);
(*BG)[ 1][ 2].ng = 3;
(*BG)[ 1][ 2].a[ 0]= 0.80000000; (*BG)[ 1][ 2].d[ 0]= 0.815902020496;
(*BG)[ 1][ 2].a[ 1]= 0.80000000; (*BG)[ 1][ 2].d[ 1]= 0.221317306997;
(*BG)[ 1][ 2].a[ 2]= 0.80000000; (*BG)[ 1][ 2].d[ 2]= 0.669619772560;
newgto H
S 4
1 13.010701000000 0.019682160277
2 1.962257200000 0.137965241943
3 0.444537960000 0.478319356737
4 0.121949620000 0.501107169599
S 4
1 13.010701000000 0.041521698254
2 1.962257200000 0.291052966991
3 0.444537960000 1.009067689709
4 0.121949620000 -1.277480448918
P 3
1 0.800000000000 0.756546169965
2 0.800000000000 0.205216749988
3 0.800000000000 0.620905772429
end
-------------------------------------------
RADIAL EXPECTATION VALUES <R**-3> TO <R**3>
-------------------------------------------
0 : 0.000000 1.965412 0.998557 1.497110 2.972853 7.305452
1 : 0.000000 2.769720 0.969842 2.227011 6.779696 23.235533
2 : 1.522453 1.066667 0.951533 1.189416 1.562500 2.230155
3 : 1.522453 1.066667 0.951533 1.189416 1.562500 2.230155
4 : 1.522453 1.066667 0.951533 1.189416 1.562500 2.230155
-------
TIMINGS
-------
Total SCF time: 0 days 0 hours 0 min 0 sec
Total time .... 0.555 sec
Sum of individual times .... 0.637 sec (114.7%)
Fock matrix formation .... 0.477 sec ( 85.9%)
Diagonalization .... 0.000 sec ( 0.1%)
Density matrix formation .... 0.000 sec ( 0.0%)
Population analysis .... 0.002 sec ( 0.4%)
Initial guess .... 0.154 sec ( 27.7%)
Orbital Transformation .... 0.000 sec ( 0.0%)
Orbital Orthonormalization .... 0.000 sec ( 0.0%)
DIIS solution .... 0.000 sec ( 0.0%)
------------------------- --------------------
FINAL SINGLE POINT ENERGY 0.072862505694
------------------------- --------------------
---------------
PLOT GENERATION
---------------
choosing x-range = -7.000000 .. 7.000000
choosing y-range = -7.000000 .. 7.000000
choosing z-range = -7.000000 .. 7.000000
GBW-File ... h.gbw
PlotType ... MO-PLOT
MO/Operator ... 1 0
Output file ... H-1.cube
Format ... Gaussian-Cube
Resolution ... 40 40 40
Boundaries ... -7.000000 7.000000 (x direction)
-7.000000 7.000000 (y direction)
-7.000000 7.000000 (z direction)
choosing x-range = -7.000000 .. 7.000000
choosing y-range = -7.000000 .. 7.000000
choosing z-range = -7.000000 .. 7.000000
GBW-File ... h.gbw
PlotType ... MO-PLOT
MO/Operator ... 2 0
Output file ... H-2.cube
Format ... Gaussian-Cube
Resolution ... 40 40 40
Boundaries ... -7.000000 7.000000 (x direction)
-7.000000 7.000000 (y direction)
-7.000000 7.000000 (z direction)
choosing x-range = -7.000000 .. 7.000000
choosing y-range = -7.000000 .. 7.000000
choosing z-range = -7.000000 .. 7.000000
GBW-File ... h.gbw
PlotType ... MO-PLOT
MO/Operator ... 3 0
Output file ... H-3.cube
Format ... Gaussian-Cube
Resolution ... 40 40 40
Boundaries ... -7.000000 7.000000 (x direction)
-7.000000 7.000000 (y direction)
-7.000000 7.000000 (z direction)
choosing x-range = -7.000000 .. 7.000000
choosing y-range = -7.000000 .. 7.000000
choosing z-range = -7.000000 .. 7.000000
GBW-File ... h.gbw
PlotType ... MO-PLOT
MO/Operator ... 4 0
Output file ... H-4.cube
Format ... Gaussian-Cube
Resolution ... 40 40 40
Boundaries ... -7.000000 7.000000 (x direction)
-7.000000 7.000000 (y direction)
-7.000000 7.000000 (z direction)
***************************************
* ORCA property calculations *
***************************************
---------------------
Active property flags
---------------------
(+) Dipole Moment
------------------------------------------------------------------------------
ORCA ELECTRIC PROPERTIES CALCULATION
------------------------------------------------------------------------------
Dipole Moment Calculation ... on
Quadrupole Moment Calculation ... off
Polarizability Calculation ... off
GBWName ... h.gbw
Electron density file ... h.scfp.tmp
-------------
DIPOLE MOMENT
-------------
X Y Z
Electronic contribution: -0.00000 0.00000 -0.00000
Nuclear contribution : 0.00000 0.00000 0.00000
-----------------------------------------
Total Dipole Moment : -0.00000 0.00000 -0.00000
-----------------------------------------
Magnitude (a.u.) : 0.00000
Magnitude (Debye) : 0.00000
Timings for individual modules:
Sum of individual times ... 4.879 sec (= 0.081 min)
GTO integral calculation ... 1.199 sec (= 0.020 min) 24.6 %
SCF iterations ... 0.733 sec (= 0.012 min) 15.0 %
Orbital/Density plot generation ... 2.947 sec (= 0.049 min) 60.4 %
****ORCA TERMINATED NORMALLY****
TOTAL RUN TIME: 0 days 0 hours 0 minutes 5 seconds 202 msec