Task 1
Explored tRNA: 1ffy
I used the "einverted" program from the EMBOSS package — search for inverted regions in nucleotide sequences.
Einverted parameters: Gap penalty 8, Minimum score threshold 0, Match score 3, Mismatch score -4
Results
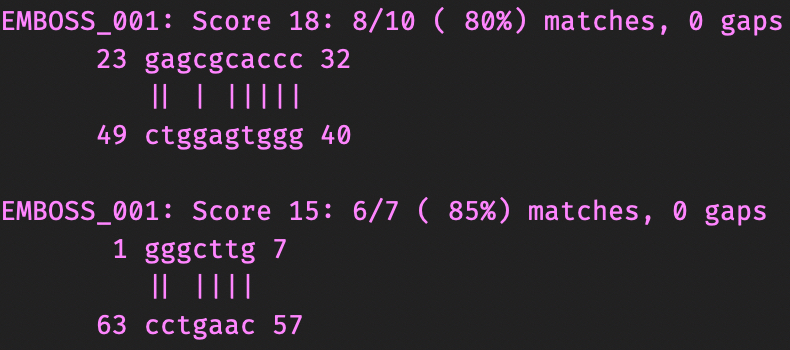
Compare with description obtained earlier using find_pair.
| Area of the structure | Positions in the structure (find_pair) | Prediction results with einverted | Prediction results with Zuker algorithm |
|---|---|---|---|
| Acceptor stem | 1-7 66-72 | 1-7 57-63 | 1-7 67-73 |
| D-stem | 10-13 22-25 | — | — |
| T-stem | 49-53 61-65 | — | — |
| Anticodon stem | 26-32 38-44 | 23-32 40-49 | — |
| Total number of canonical nucleotides pairs | 20 | 14 | 27 |
I'm not sure why, but einverted and Zuker algorithm performed poorly with the task.
Task 2. Search for DNA-protein contacts in a given structure
Ex.1
Links to scripts:
script1 script2Ex.2
script3| Contacts of protein atoms with | Polar | Non-polar | Total |
|---|---|---|---|
| 2'-deoxyribose residues | 1 | 20 | 21 |
| phosphoric acid residues | 13 | 60 | 73 |
| residues of nitrogenous bases from the side of the major groove | 4 | 9 | 13 |
| residues of nitrogenous bases from the side of the minor groove | 0 | 1 | 1 |
Contacts between DNA and non-polar atoms (C, P, S) of the protein are the most often countered
Ex.3
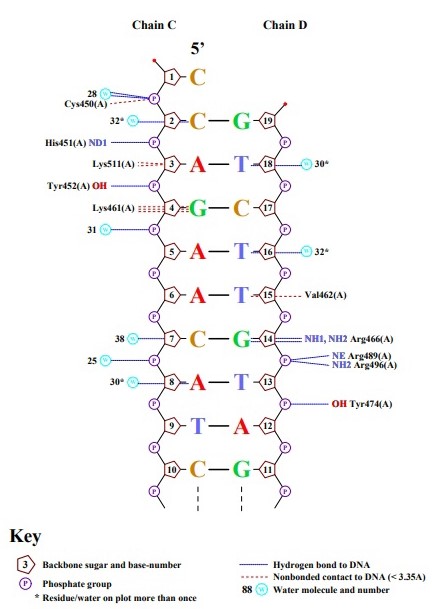
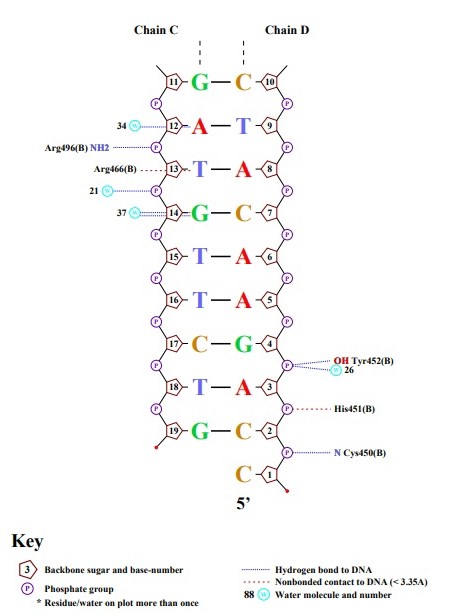
Ex.4
Lys461 has the largest number of contacts with DNA — 3, but I don't know where they are
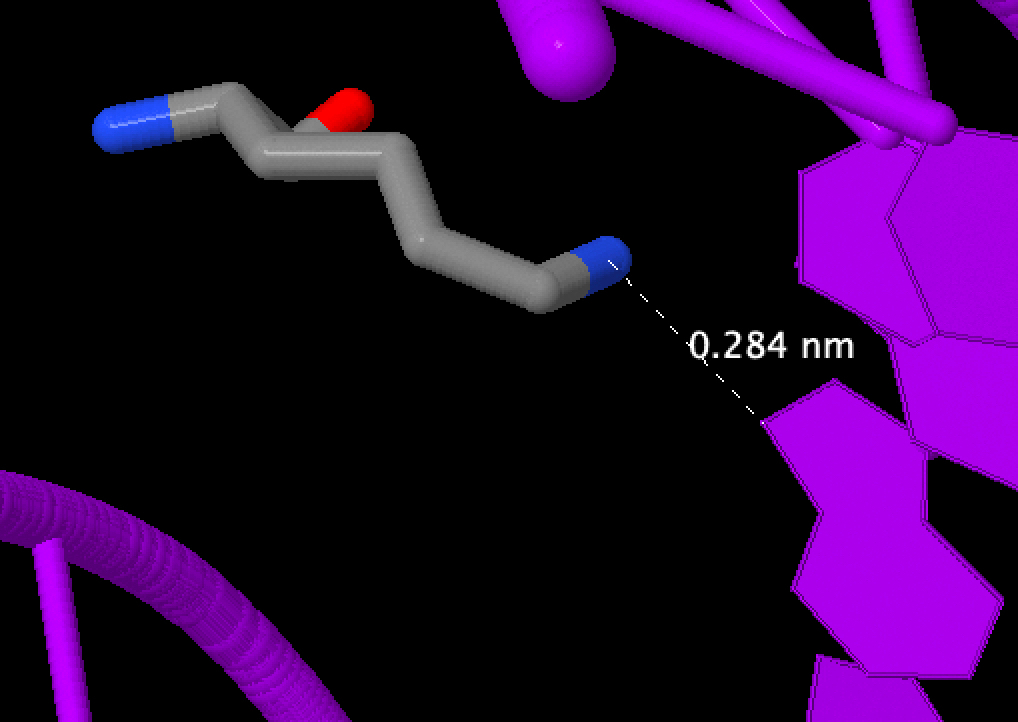
Besides Lys461, Val462 should be important for DNA sequence recognition, because they both recognise nitrogenous bases
But how valine can interact with thymine remains a mystery. Troubles with the nucplot or with me?