Mini-review
2023, Moscow
Mini-review
Genome and proteome overview of archaea Haloprofundus salinisoli
Tarasov Alexey
Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University
E-Mail: Geonosianin@fbb.msu.ru
Annotation
This mini-review is devoted to the study of the genome and proteome of the archaea Haloprofundus salinisoli. The work uses the functionality of spreadsheets and programming in Python. The research of the mini-review contributes to the study of the genome of halophilic archaea and creates the ground for further experiments in this sphere.
I. Introduction
Taxonomy:
- Domain: Archaea
- Phylum: Methanobacteriota
- Class: Halobacteria
- Order: Haloferacales
- Family: Haloferacaceae
- Genus: Haloprofundus
- Specie: Haloprofundus salinisoli
Habitat:
Haloprofundus salinisoli are extremophiles that live in the salt lakes and saline soils. This halophilic archaea has a wide diversity in the western region of China.
Features:
The name of the species comes from the words “salinus” (salty) and “solum” (soil). Cells are stain Gram-negative. Distilled water leads to cell lysis - the minimum concentration of salt in the water to prevent lysis and support vital activity should be at least 12%. The following substrates may be used by archaea as carbon and energy sources for growth: d-mannose, glycerol, d-mannitol, d-sorbitol, acetate, pyruvate, dl-lactate, succinate, l-malate, fumarate, l-glutamate and citrate. In this work, the genome and proteome of the archaea Haloprofundus salinisoli are investigated. Based on the table (taken from ftp.ncbi.nlm.nih.gov) a histogram of protein lengths was constructed, intergenic gaps and intersecting protein genes have been researched. Moreover, the start and stop codons in the genome of Haloprofundus salinisoli have been determined and their quantitative ratio has been established [1].
II. Materials and Methods
Data on the genome of the studied bacterium were taken from the official websites of the NCBI (National Center for Biotechnology Information) and ResearchGate. Google Sheets spreadsheets were used to analyze the received data. The program was written in the Python programming language.
III. Results and Discussion
Statistic data about the genome and the proteome
Protein lengths
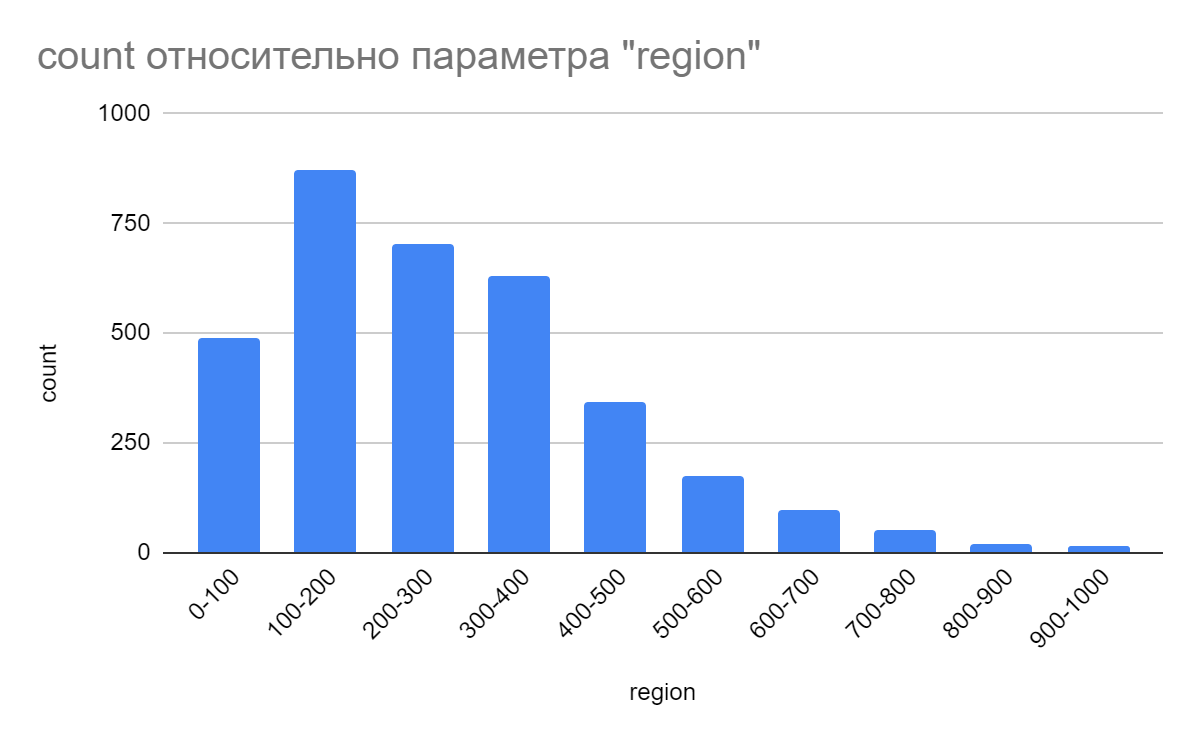
The resulting histogram of the proteome reflects the number of proteins and their length in the archaea proteome (S2 in supplementary material). It follows from the histogram that the largest number of proteins that make up the bacterial proteome are 200-300 amino acids long.
This corresponds to the average protein length of 283 amino acids proved for all archaea [2].
The length of 0-100 and 200-400 amino acids has a smaller number of proteins. A small part of proteins reaches the size of 900-1000 amino acids. Also, the histogram shows that only a small part of the total number of proteins reaches a length of more than 500 amino acids [4].
The size of several proteins is significantly larger than the rest in size. This may indicate a specific function performed by such proteins.
The size of a protein can affect several properties and functions of a protein. In theory, such proteins should have greater stability, since they have more opportunities for proper laying and structure formation and high affinity for ligands (molecules with which they bind), which can lead to higher efficiency of their functioning.
Lengths of the gaps between the genes of proteins
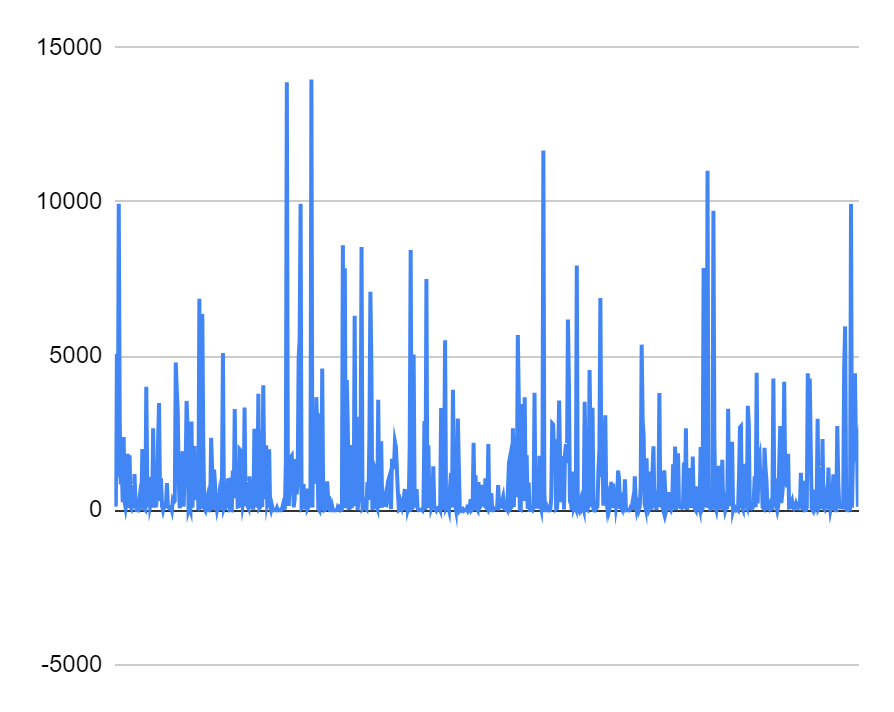
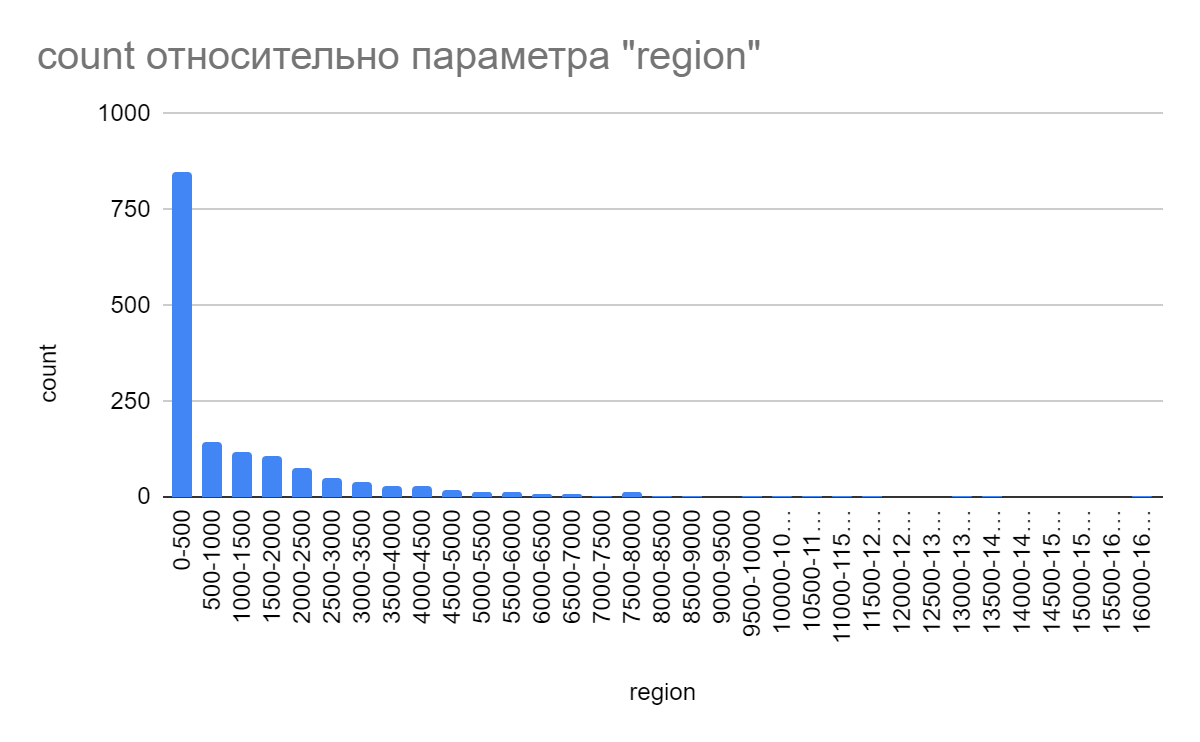
A study of data on the gaps between genes was conducted, as a result of which it became possible to construct a histogram of the distances between coding sequences (CDS) on the plus chain of the largest chromosome, as well as a diagram of the distribution of distances (S2 in supplementary material). According to the presented diagram, the vast majority of the intergenic gaps have a length from 0 to 500 pairs of nucleotides. At the same time, a small part of the distances between genes reaches a length of more than 5,000 pairs of nucleotides. The maximum length of the gap recorded in the table was 16264 pairs of nucleotides. The average length of the gaps is approximately 1248. The median value is 199 pairs of nucleotides [4].
Number of nucleotides in intersection
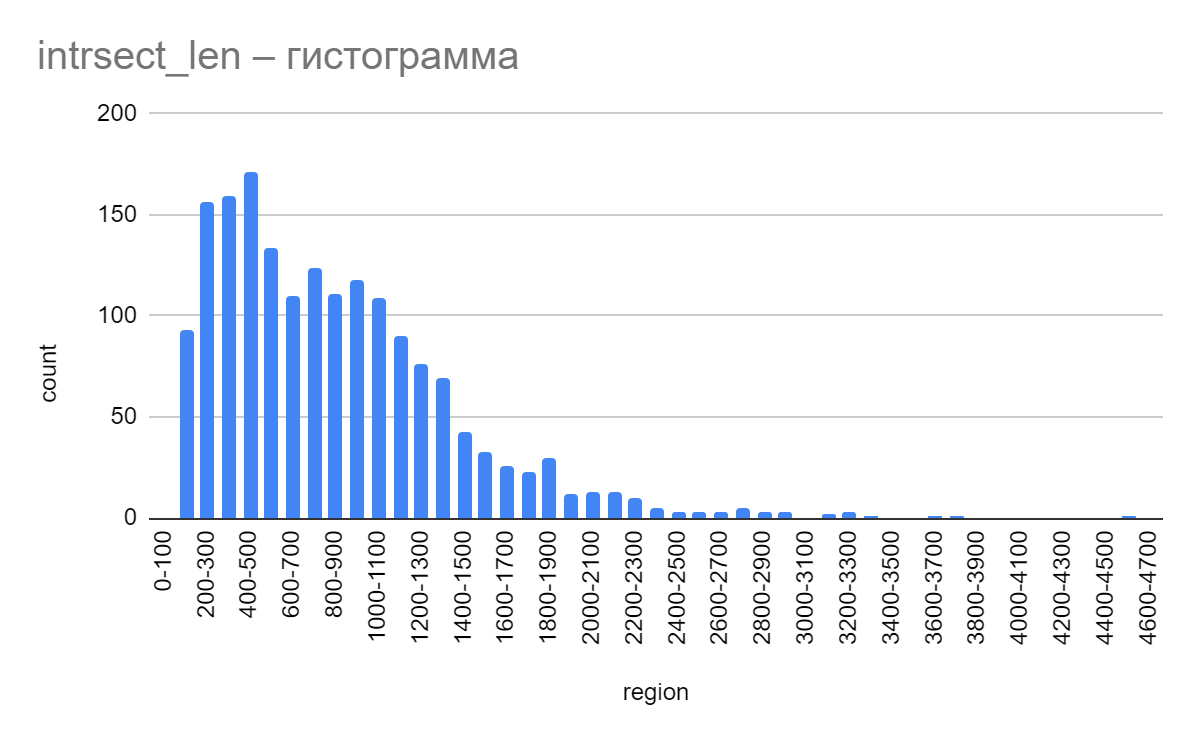
A study of data on intersecting genes of protein sequences was conducted, as a result of which it became possible to construct a histogram of the number of nucleotides in the intersection in coding sequences (CDS) on the plus chain of the largest chromosome (S2 in supplementary material). From the data obtained it follows that аll of the coding sequences (СDS) on the plus chain of the largest chromosome intersect with the next CDS on the plus chain. According to the presented histogram, the number of nucleotides in the intersection is most often from 200 to 1100. The most common number is 400-500 nucleotides. At the same time, an insignificant part of the sequences contains more than 4500-4600 intersecting nucleotides. The maximum number recorded in the table was 4535 nucleotides at the intersection. The average number of nucleotides is approximately 841. The median value is 749 nucleotides [4].
The number of gene intersections can affect the complexity and functionality of the genetic code. A large number of intersecting genes contribute to the interaction of these genes and the formation of complex regulatory networks. Most likely, a large number of genes in the intersection may indicate that these sections of the sequence ensure the flow of complex processes in the organism of the archaea that require regulation by several genes at once.
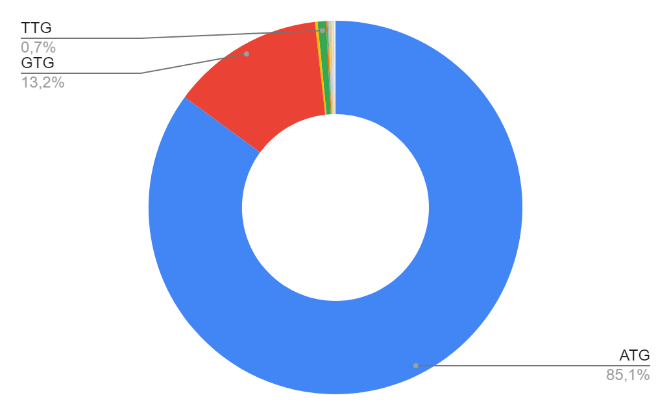
Start and stop codons
Using a fasta file and the Python programming language, a study was conducted to determine the start and stop codons of protein genes, as well as to identify the frequency of their occurrence. The program reads the fasta file and outputs, separated by a space, the name of the start (stop) codon and the number of times it occurred in the genome. The results are presented in a table and in a diagram. (S1 in supplementary material)
| Start codon | Frequency | Start codon | Frequency | Start codon | Frequency |
| ATG | 2942 | GGG | 1 | ATA | 2 |
| GTG | 456 | ATC | 3 | TCC | 2 |
| CTG | 8 | GAC | 1 | GAA | 2 |
| TTG | 25 | CGT | 1 | GCC | 1 |
| ATT | 4 | GGC | 1 | AAG | 2 |
| CAG | 2 | GCG | 2 | TGG | 1 |
| ACC | 1 | ACT | 1 | TCT | 1 |
| STOP CODON | FREQUENCY | STOP CODON | FREQUENCY |
| TGA | 1975 | GAA | 1 |
| TAA | 745 | GTG | 1 |
| TAG | 724 | TCC | 2 |
| GAT | 2 | GGA | 2 |
| CGG | 1 | GAG | 2 |
| TAC | 1 | ATC | 1 |
| GAC | 1 | GCG | 1 |
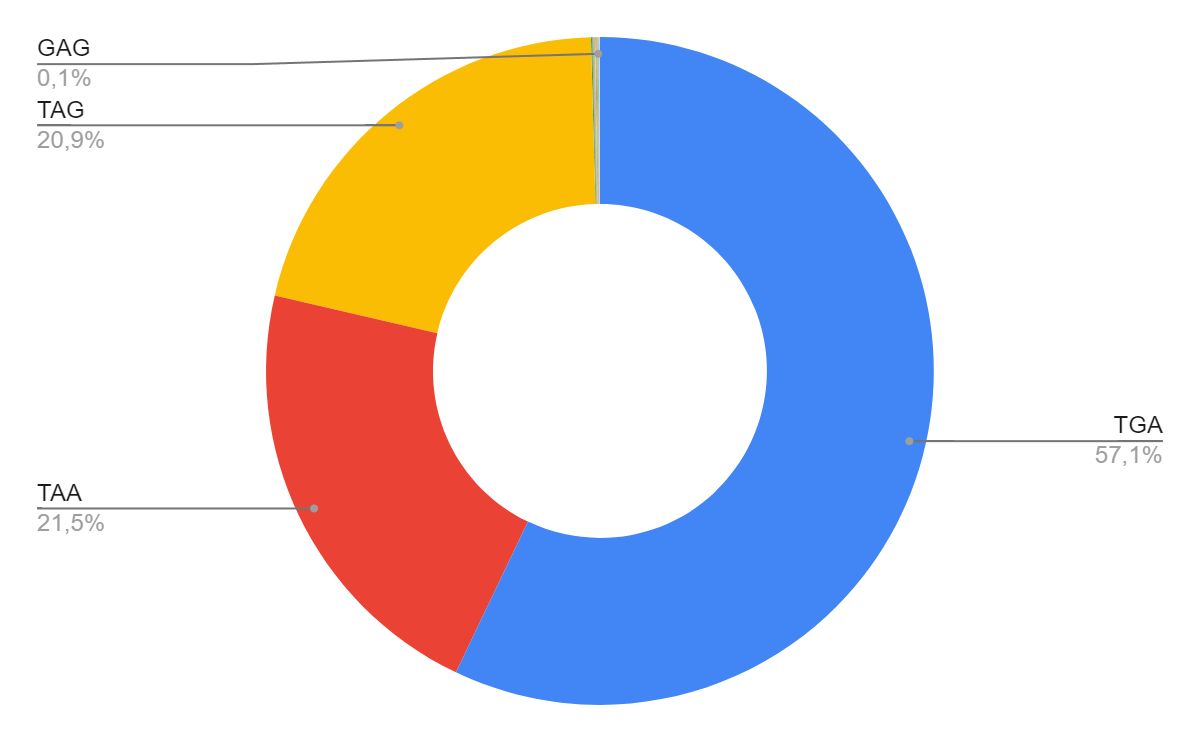
This data reveals that ATG has become the most common start codon of protein genes. The frequency of its occurrence is 2942 times, which is about 85.1% of the total number of start codons in the sequence. ATG (AUG) is the most common start codon among living organisms. The most common stop codon of protein genes was TGA. The frequency of its occurrence is 1975 times, which is about 57.1% of the total number of start codons in the sequence. Stop codons TAA and TAG are also common (the frequency of occurrence is 745 and 724, respectively, which is about 42% of all stop codons of protein sequences). TGA, TAA and TAG (UGA, UGA and UAG) are the most common stop codons among living organisms.
It is obvious from the results that the ATG start codon is characteristic of this type of archaea, but GTG and less often TTG are also often used, which differ from it by only one nucleotide. These two codons are probably similar to ATG due to the slight difference between them.
The rest of the atypical codons are often the start codons of pseudogenes. Therefore, it can be assumed that a change in the start codon plays a role in the shutdown of genes. It is also possible that such genes have changed or lost their functions because of a mutation [3].
There are also many atypical stop codons. Their presence in the genome can be explained by mutation or error in sequencing. It is likely that such stop codons can perform additional functions, for example, encode an amino acid [5][6].
Conclusion
Despite the fact that archaea represent a very important group in the evolution of living organisms on Earth, in general they remain insufficiently studied. Obviously, in order to get answers to many questions about their existence and development, further study of archaea is required.
References:
- Si‑Ya Li1, Yu‑Jie Xin1, Chen‑Xi Bao1, Jing Hou1, Heng‑Lin Cui. (2022) Haloprofundus salilacus sp. nov., Haloprofundus halobius sp. nov. and Haloprofundus salinisoli sp. nov.: three extremely halophilic archaea isolated from salt lake and saline soil.
- Tiessen, A., Pérez-Rodríguez, P. & Delaye-Arredondo, L.J. (2012) Mathematical modeling and comparison of protein size distribution in different plant, animal, fungal and microbial species reveals a negative correlation between protein size and protein number, thus providing insight into the evolution of proteomes. BMC Res Notes 5, 85.
- Ariel Hecht, Jeff Glasgow, Paul R. Jaschke, Lukmaan A. Bawazer, Matthew S. Munson, Jennifer R. Cochran, Drew Endy, Marc Salit (21.02.2017). "Measurements of translation initiation from all 64 codons in E. coli". Nucleic Acids Research (Vol. 45, Iss. 7, P. 3615–3626)
- National Center for Biotechnological Information.
- James H. Campbell, Patrick O’Donoghue, Alisha G. Campbell, Patrick Schwientek, Alexander Sczyrba, Tanja Woyke, Dieter Söll and Mircea Podar. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci U S A. (2013 Apr 2) 110(14): 5540–5545.
- Наталья Резник. Неоднозначность стоп-кодонов. «Троицкий вариант» №24(218), 6 декабря 2016 года