Bioinformatics
Mini Review of Lactobacillus amylovorus GRL1118
Polteva Izabella
Moscow State University M. V. Lomonosov Department of Bioengineering and bioinformatics
Contacts: ibavpa@gmail.com
________________________________________________
Introduction
A big complex mixture of bacteria has colonized the gastrointestinal tract of porcine. It performs various functions that are good to the host. Unlike in humans, lactobacilli are the dominant lactic acid bacteria in the pig intestine (Ravi Kant, Lars Paulin, Edward Alatalo, Willem M de Vos, Airi Palva. Genome sequence of Lactobacillus amylovorus GRL1118, isolated from pig ileum. Journal of Microbiology Vol. 193, No. 12 (26 may 2011)).
The taxonomy is presented in table 1
Tab 1. Taxonomy of L. amylovorus GRL1118
| Superkingdom | Bacteria |
| Phylum | Bacillota |
| Class | Bacilli |
| Order | Lactobacillales |
| Family | Lactobacillaceae |
| Genus | Lactobacillus |
| Species | Lactobacillus amylovorus |
| Strain | Lactobacillus amylovorus GRL1118 |
L. amylovorus GRL1118 is a Gram-positive, anaerobic, rod-shaped bacterium that was isolated from porcine ileum. It has typical features of homofermentative Lactobacillus species, like the production of large quantities of lactic acid and small amounts of acetic acid but no gas from glucose.
In this research the genome of Lactobacillus amylovorus GRL1118 will be investigated.
Methods
The data of Lactobacillus amylovorus GRL1118 and Lactobacillus amylovorus GRL1112 have been taken from website NCBI (National Center of Biotechnology Information). For counting of nucleotide composition of the genome Python and Terminal (Bash) had been used. For processing information Google Sheets had been used.
Results
1. Proteome data
Proteome of Lactobacillus amilovorus GRL1118 consists of 1 chromosome and 2 plasmids that are coding a few types of RNA (tRNA, rRNA, ncRNA, tmRNA). The number of transfer RNA (tRNA) genes is 62. This is 80.5% of all RNA genes in the genome. Number of ribosomal RNA genes (rRNA) on the chromosome is 12, this is 15.6% of all RNA genes. There are 2 genes of ncRNA (non-coding RNA) (2.6%) and 1 tmRNA (transfer-messenger RNA) (1.3%)
Tab 2. Number of types of RNA in genome of L. amylovorus GRL1118
| features | tRNA | rRNA | ncRNA | tmRNA | CDS |
| chromosome | 62 | 12 | 2 | 1 | 1851 |
| plasmid 1 | 0 | 0 | 0 | 0 | 1672 |
| plasmid 2 | 0 | 0 | 0 | 0 | 75 |
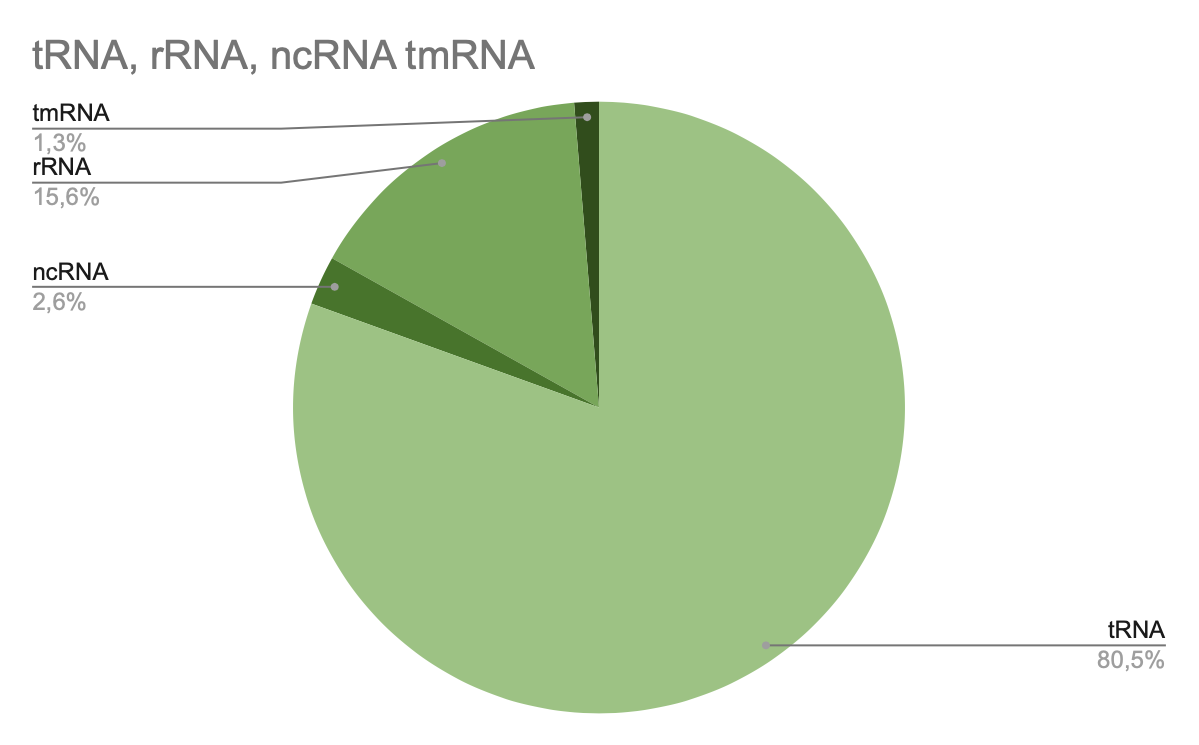
Pic 1. L. amylovorus GRL1118 ratio RNA pie-chart
2. Comparing bacteria
Ratios of types of RNA of Lactobacillus amylovorus GRL1118 and Lactobacillus amylovorus GRL1112 have also been compared
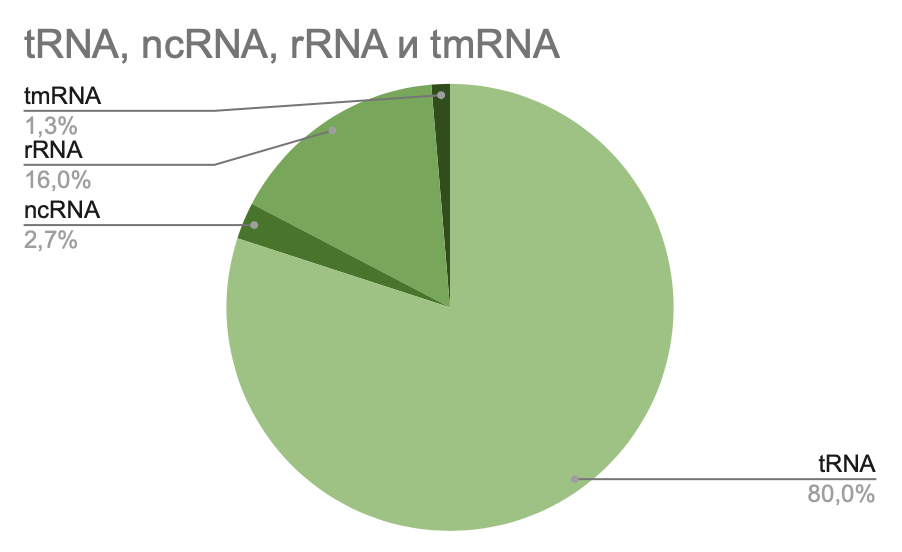
Pic 2. L. amylovorus GRL1112 ratio RNA pie-chart
The pie-chart shows that there are four types of RNA in the genome of Lactobacillus amylovorus GRL1118 as in Lactobacillus amylovorus GRL1118. The number of identical types of RNA coincides, and this improve that these bacteria belong to one species
Tab 3.Number of types of RNA in genome of L. amylovorus GRL1112
| features | tRNA | rRNA | ncRNA | tmRNA | CDS |
| chromosome | 60 | 2 | 12 | 1 | 2062 |
| plasmid 1 | 0 | 0 | 0 | 0 | 37 |
| 0 | 0 | 0 | 0 | 0 | 28 |
3. Protein length histogram
Based on the data from the table (Protein length histogram), a histogram was created. It displays the number of protein length distributions in a Lactobacillus amylovorus GRL1118:
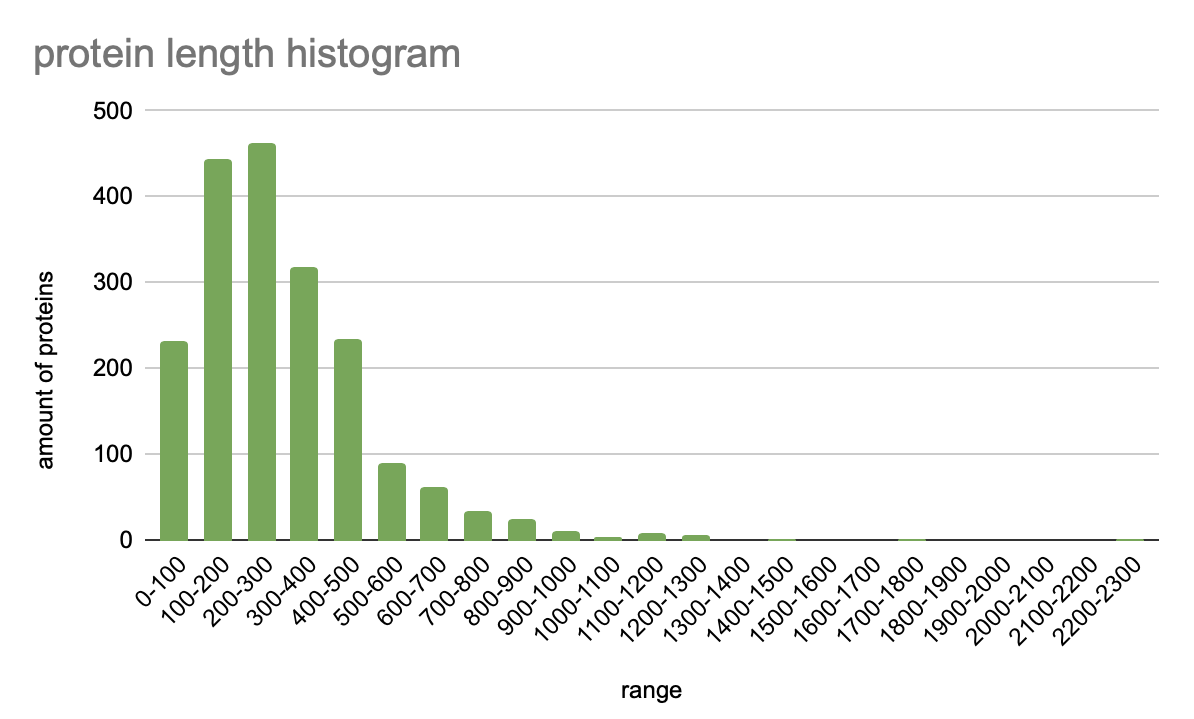
Pic 3. Protein length histogram
On this histogram you can see not a large variety of lengths. The biggest amount of those proteins which length is in the range from 200 to 300 amino acids. In second place are proteins whose length 100-200 amino acids. After CDSs with 400 amino acids, the number of proteins decreases sharply, which can be seen in the histogram. There are only two proteins with length 1400-1500. And there are no proteins whose length is in the range from 1500-1700 and 1900- 2200 amino acids.
4. GC% content in genome
Based on the data from the table, a histogram was compiled that shows the percentage of guanine and cytosine in the coding sequences.
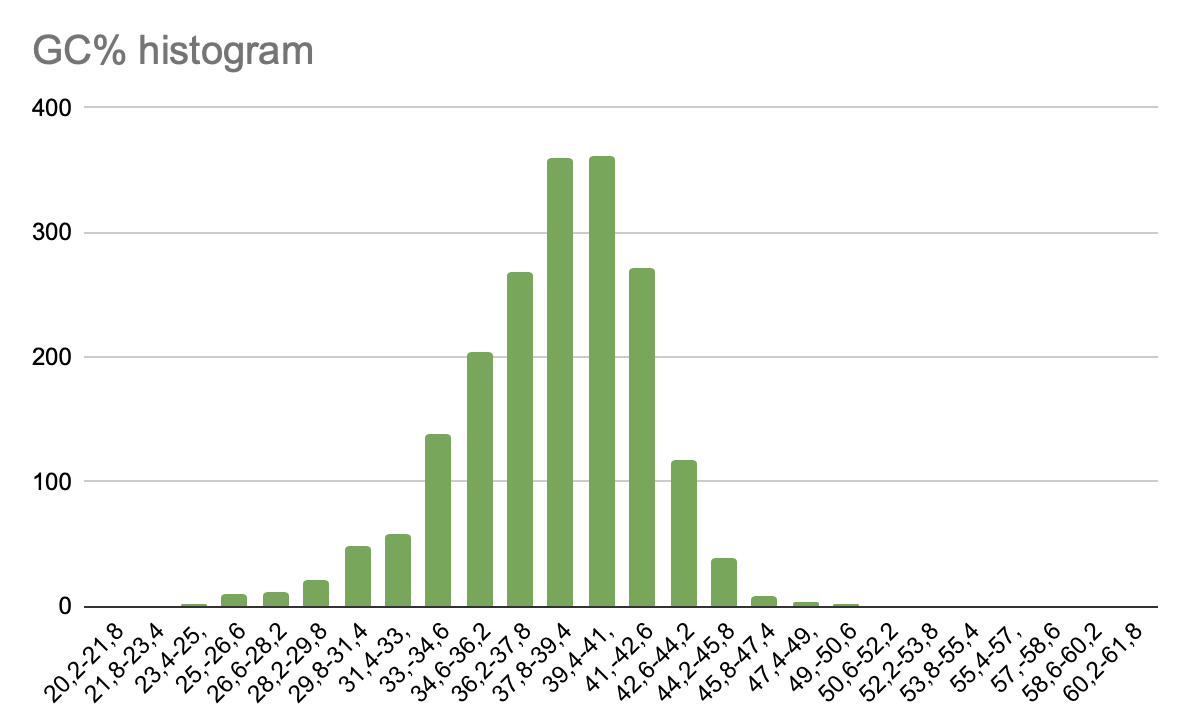
Pic 4. GC% histogram in CDS
On this histogram we can see that the largest number of CDS are those that contain 37.8-39.4 and 39.4-41.0 percent of guanine and cytosine. If we move along the x-axis in opposite directions from these values, we can see that the diagram decreases. The smallest number of CDS are those that contain 20.2-25.0 and 49.0-60.63 percent of guanine and cytosine.
5. Nucleotides % in replicones
During the studying the genome of Lactobacillus amylovorus GRL1118, nucleotide composition of replicons has been investigated. Based on this data the pie-chart of nucleotide composition in each replicon has been made.
a. Chromosome
On this diagram we can see that most adenine is most abundant in chromosomes. It takes 35.1% of the total amount (588820). In the second place is thymine, it takes 30,8% (582911). There are only 18.7% of cytosine in chromosome (354694).
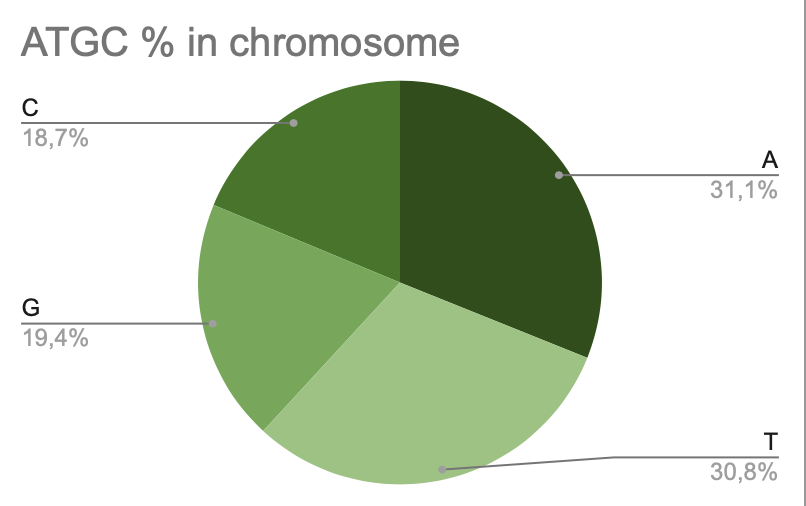
Pic 5. ATGC% in chromosome of Lactobacillus amylovorus
b. Plasmid 1
It is seen that adenine is the most common in plasmid 1, 35,1% (1594). A bit less contains thymine, about 30,7% (1393). And only 15,9% guanine is in plasmid 1 (721).
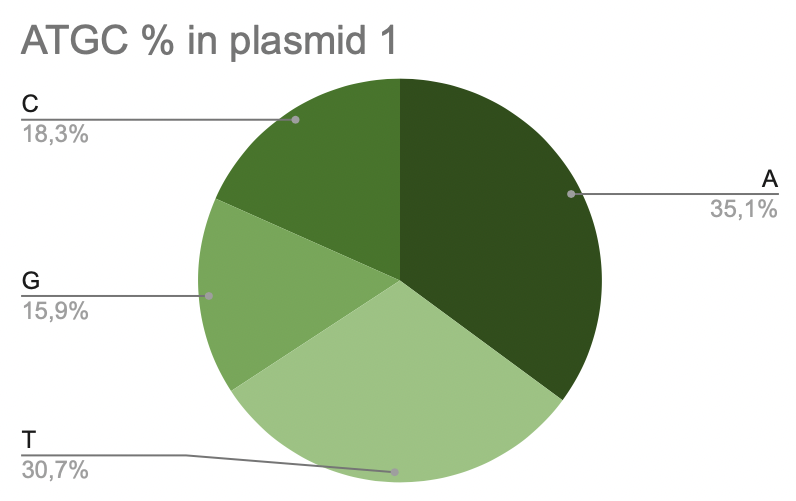
Pic 6. ATGC% in plasmide 1 of Lactobacillus amylovorus GRL1118
c. Plasmid 2
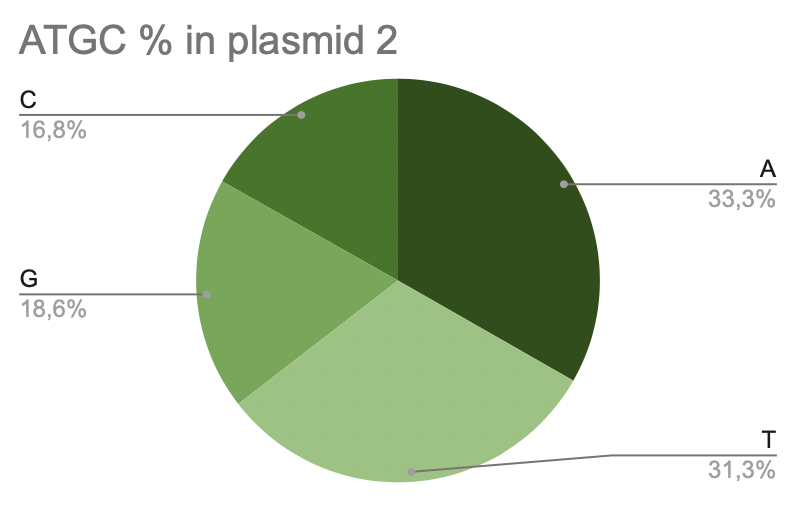
Pic 7. ATGC% in plasmide 2 of Lactobacillus amylovorus GRL1118
Most of all is adenine in plasmid 2, it takes 33,3% from the total quantity (25988). Plasmid 2 contains approximately the same amount of thymine, 31,3% (24436). And it contains the least amount of cytosine, only 16,8% (13148).
6. Stop-codons
During the investigation of the genome of Lactobacillus amylovorus GRL1118, stop-codon composition has been found. Using this data the diagram of percentage has been made.
a. Chromosome
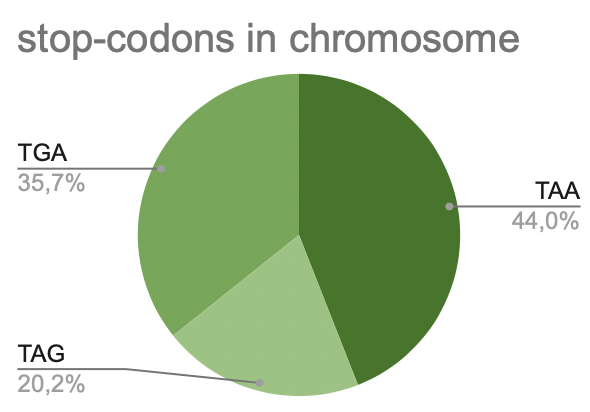
Pic 8. stop-codons in chromosome
There are 64 codons, and three of them are called stop-codons. When the ribosome reaches these codons, the translation stops. These codons are TAA, TAG, TGA. It is seen that most of all is TAA. It takes 44%. On the second place is TGA. It takes 35,7%. The rest is occupied by TGA (20,2%).
b. Plasmid 1
On this histogram we can see, most of it is taken up by TAA (45.7%). Less of it is taken by TAG (22.5%). 31.8% is taken by TGA.
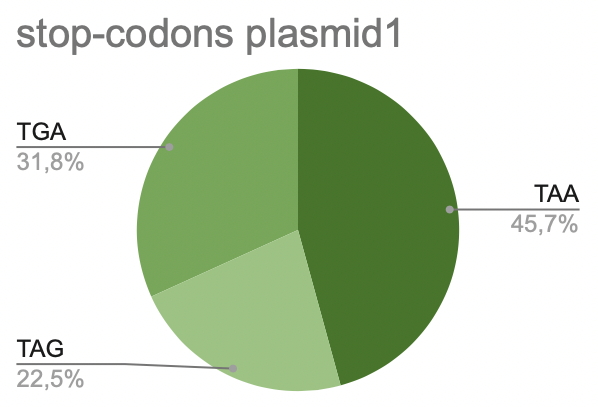
Pic 9. stop-codons in plasmid 1
c. Plasmid 2
It is seen that most of all is taken by TAA (45.7%). TGA takes about 31.8%. Less of all is taken by TAG (22.5%).
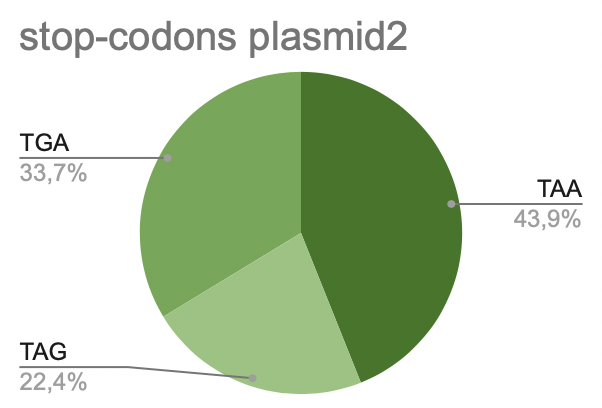
Pic 10. stop-codons in plasmid 2
Literature
1. Description of Lactobacillus amylovorus GRL1118: Ravi Kant, Lars Paulin, Edward Alatalo, Willem M de Vos, Airi Palva. Genome sequence of Lactobacillus amylovorus GRL1118, isolated from pig ileum. Journal of Microbiology Vol. 193, No. 12 (26 may 2011)
https://journals.asm.org/doi/10.1128/jb.00423-112. Ulla Hynönen, Ravi Kant,, Tanja Lähteinen, Taija E Pietilä, Jasna Beganović, Hauke Smidt, Ksenija Uroić, Silja Åvall-Jääskeläinen and Airi Palva. Functional characterization of probiotic surface layer protein-carrying Lactobacillus amylovorus strains. BMC microbiology Article number: 199 (28 July 2014)
https://bmcmicrobiol.biomedcentral.com/articles/10.1186/1 471-2180-14-1993. Taxonomy of Lactobacillus amylovorus GRL1118
https://www.ncbi.nlm.nih.gov/datasets/taxonomy/tree/?taxo n=6955624. Lactobacillus amylovorus GRL1118 genome data
https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/194/ 115/GCF_000194115.1_ASM19411v15. Lactobacillus amylovorus GRL1112 genome data
https://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/182/855/ GCF_000182855.2_ASM18285v1/Accompanying materials
1. Proteome data
https://docs.google.com/spreadsheets/d/1R0USTsd6HvrZ1 HCJCMjNIed4eJ0fJXbRdKZIpzknKxo/edit?gid=1674958449 #gid=16749584492. Types of RNA in genome of Thermococcus profundus
https://docs.google.com/spreadsheets/d/1R0USTsd6HvrZ1 HCJCMjNIed4eJ0fJXbRdKZIpzknKxo/edit?gid=1392123172 #gid=13921231723. Protein length histogram
https://docs.google.com/spreadsheets/d/1Vcitd3IGjmQMrS DT7FpzYEsxTQdu4PzJ6XqNbfrzR1A/edit?gid=2112773930 #gid=21127739304. GC% in replicons
https://docs.google.com/spreadsheets/d/1Vcitd3IGjmQMrS DT7FpzYEsxTQdu4PzJ6XqNbfrzR1A/edit?gid=667603809# gid=6676038095. Stop-codons
https://docs.google.com/spreadsheets/d/1R0USTsd6HvrZ1 HCJCMjNIed4eJ0fJXbRdKZIpzknKxo/edit?gid=160283876# gid=1602838766. Code
https://drive.google.com/file/d/1fZTUX4QAX8XabtVq3ErZX7 TqNtsRSvce/view