Bioinformatics, 2023
Advance Access Publication Date: DD december 2023
Minireview
Genome and proteome analysis of archaeon Succinvibrio dextrinosolvens
Chistiakova Ekaterina
Department of Bioengineering and Bioinformatics, Lomonosov Moscow State University, Moscow, Russia
Abstract: Archaea have been shown to naturally produce or can be engineered to produce a range of products such as biofuels (e.g., biomethane, biohydrogen, bioethanol, or biobutanol), bioplastics (PHA), compatible solutes, nanobiotechnology components (surface-layer proteins, lipids) and precursor chemicals (e.g., acetate, 2,3-butanediol) needed for the industrial synthesis of high value chemicals. In order to take advantage of these remarkable features of the bacterial genome, it is necessary to study its structure. In this mini-review we discuss what features can be found in the genome of a particular archaeon.
Contact: katyach@fbb.msu.ru
1. Introduction
Succinivibrio dextrinosolvens is an anaerobic, gram-positive organism isolated from ruminant animals and belongs to the phylum Firmicutes. This microorganism is capable of fermenting various carbohydrates, including dextrin, glucose, and other sugars, producing succinate, acetate, formate, hydrogen, and carbon dioxide[5].
Succinivibrio dextrinosolvens is of significant importance in the digestive system of ruminant animals, as these bacteria inhabit the rumen and play a role in food processing and metabolism. They participate in substrate metabolism and utilization, thereby playing a key role in the digestion of ruminant animals. The appearance of the archaeon is illustrated on the Fig.1.
Domain "Bacteria"
Phylum Pseudomonadota
Class Gammaproteobxacteria
Order Aeromonadales
Family Succinivibrionaceae
Genus Succinivibrio
Species Succinivibrio dextrinosolvens
2. Methods
3.1 Data on the genome
Succinvibrio dextrinosolvens has 2 ring chromosomes with the unequal distribution of genomic data. (Table.1) In total, genome encodes 2476 genes, 2382 CDSs, 69 tRNA, 21 rRNA, 3 ncRNA, 1 tmRNA.
It can be concluded that the second ring DNA is evolutionarily more recent, since most of the information about the genome is contained in the first chain. In future studies, it will be interesting to find out what will change if cut out the second DNA chain.Table.1.
| genomic accession | seq type | chromosome | CDS | ncRNA | rRNA | tmRNA | tRNA |
|---|---|---|---|---|---|---|---|
| NZ_CP068345.1 | chromosome | 1 | 2222 | 69 | 21 | 3 | 1 |
| NZ_CP068346.1 | chromosome | 2 | 160 | 0 | 0 | 0 | 0 |
3.2 Protein length data
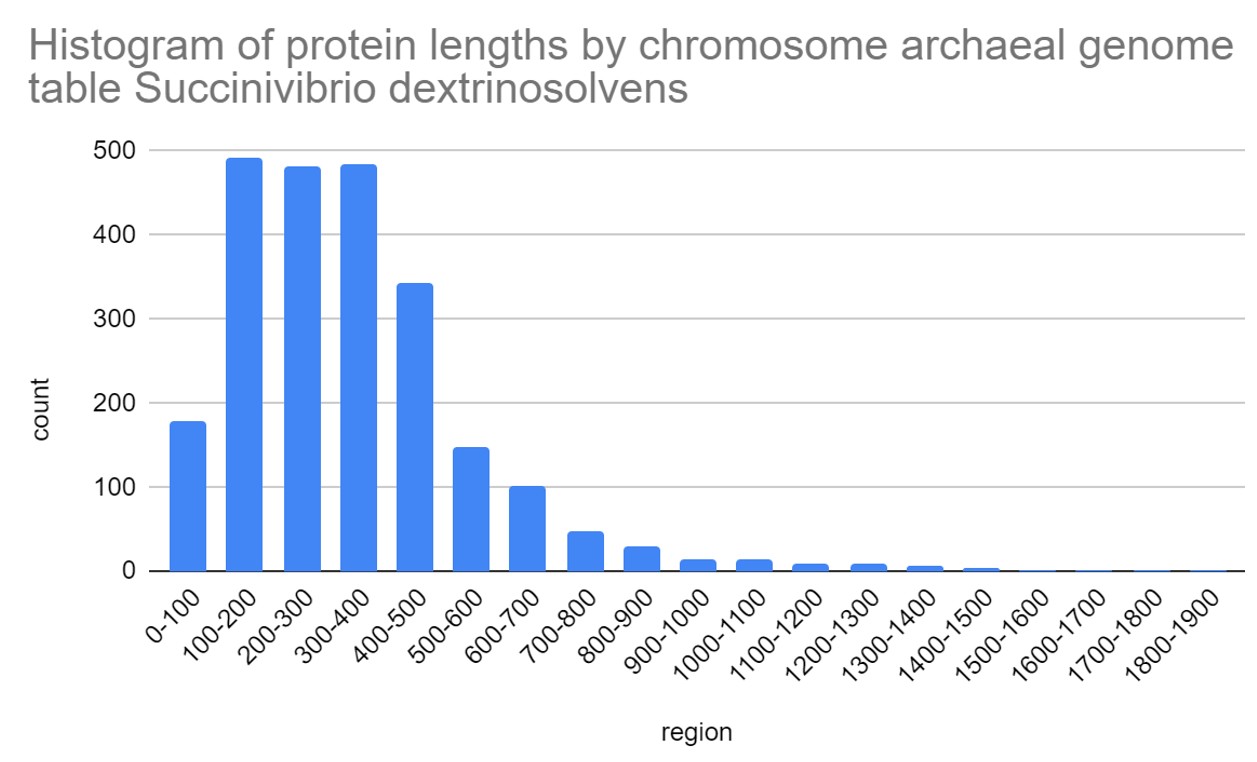 Fig.2. Histogram of protein length by chromosome archaeal genome table Succinivibrio dextrinosolvens.[4]
Fig.2. Histogram of protein length by chromosome archaeal genome table Succinivibrio dextrinosolvens.[4]
The genome of Succinivibrio dextrinosolvens encodes 2383 proteins. The median length of proteins is 308. The maximum length of a protein is 1961 amino acids, the minimum is 37. The distribution of protein lengths is presented in the histogram (Fig.2). By comparing this data with data from other bacteria, it can be seen that protein lengths of Succinivibrio dextrinosolvens are slightly longer than the average length of archaeal proteins (242 amino acids) according to BMC(biomedcentral)[7].
3.3 Start codons
There are 3 types of start codons in the coding sequences of the genome. Their quantities are presented in fig3. In accord with this ranking of the start codons, genes starting with ATG are, on average, expressed at significantly higher levels than genes that start with GTG, and the latter are expressed at higher levels than genes starting with TTG. ATG is the optimal start codon that is actively maintained by purifying selection. In prokaryotes, translation start signals are subject to weak but significant selection for maximization of initiation rate and, consequently, protein production[9].
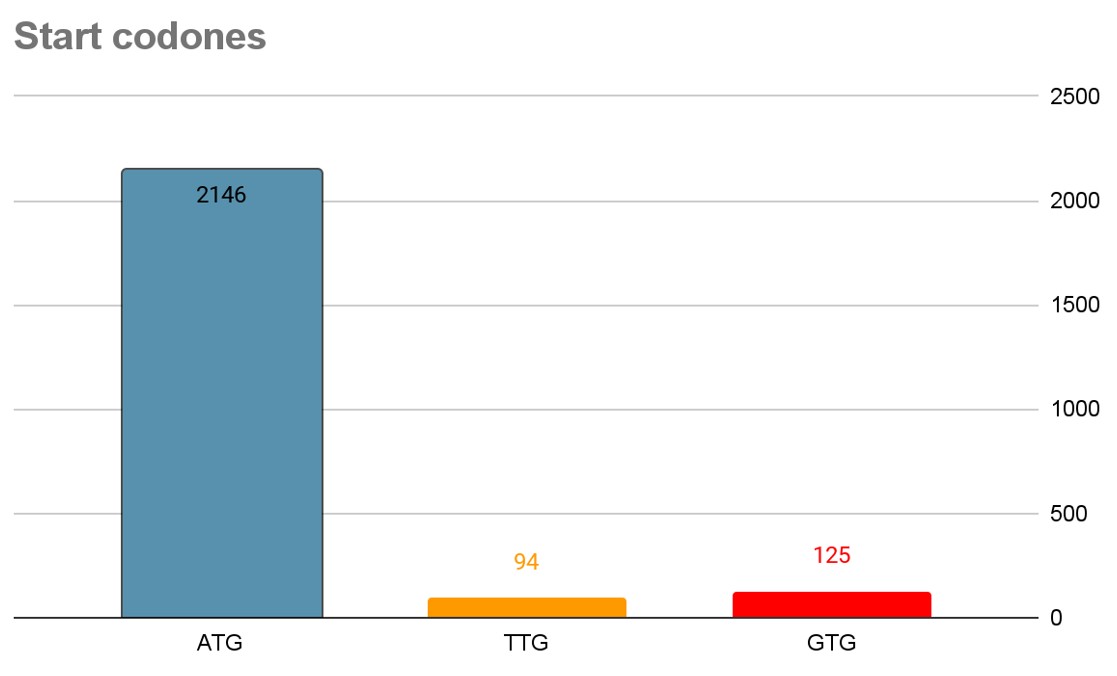
Fig.3. Bar chart of start codons
3.4 Stop codons
There are 3 types of stop codons in the coding sequences of the genome. Their quantities are presented in table.2.
Protein coding genes terminate with one of three stop codons (TAA, TGA, or TAG) that, like synonymous codons, are not employed equally.
According to the NCDI research, the frequency of TAA and TGA depends on genomic GC content (with increasing genomic GC content the frequency of the TAA codon decreases and that of the TGA codon increases in a reciprocal manner), while the incidence of TAG is irrespective of the CG content[8].
Table.2. Stop codons
| stop codons | quantity |
|---|---|
| TAA | 1465 |
| TGA | 196 |
| TAG | 655 |
From the data obtained we can conclude that the GC content in the genome is quite low (the frequency of TAA is much higher than of TGA).
3.4 Intersecting sequences CDS
In the longest chromosome chain (1) there are 59 sequences of proteins, which intersect with the next CDS on the same chain. In this way, a nucleotide sequence may make a contribution to the function of one or more gene products. The biggest number of sequences cross the following via 3 nucleotides. Other overlap lengths are presented in Table.6.
Gene overlap can be explained by finding them in different reading frames or by alternative splicing. In this way, a nucleotide sequence may make a contribution to the function of one or more gene products.
Table.3. Intersecting sequences CDS
| length of intersection | 3 | 6 | 7 | 10 | 13 | 15 | 16 | 18 | 19 | 21 | 22 | 30 | 34 | 72 | 88 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| quantity | 15 | 4 | 18 | 2 | 5 | 2 | 3 | 2 | 1 | 1 | 2 | 1 | 1 | 1 | 1 |
4.1. SUPPLEMENTARY MATERIALS
The table contains data on the archaea genome as well as individual pages with calculations for each of the aspects presented in section 3.
4.2. SOURCES
1. Kevin Pfeifer, İpek Ergal, Martin Koller, Mirko Basen, Bernhard Schuster, Simon K.-M.R. Rittmann, Archaea Biotechnology, Research review paper, March–April 2021, 107668
2. Nelson, Chase W; "Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic", National Library of Medicine, 1 October 2020
5. Bacteremia Due to Succinivibrio dextrinosolvens Report of a Case PAUL M. SOUTHERN, JR., M.D. Department of Pathology, The University of Texas Health Science Center at Dallas, Dallas, Texas 75235:
7. Nevers, Y., Glover, N.M., Dessimoz, C. et al. Protein length distribution is remarkably uniform across the tree of life. Genome Biol 24, 135 (2023):
8. Korkmaz G, Holm M, Wiens T, Sanyal S. Comprehensive analysis of stop codon usage in bacteria and its correlation with release factor abundance. J Biol Chem. 2014 Oct 31;289(44):30334-30342. doi: 10.1074/jbc.M114.606632. Epub 2014 Sep 12. PMID: 25217634; PMCID: PMC4215218.
9. Belinky F, Rogozin IB, Koonin EV. Selection on start codons in prokaryotes and potential compensatory nucleotide substitutions. Sci Rep. 2017 Sep 29;7(1):12422.