Minireview of the genome of Ciceribacter thiooxidans
Iakov Korobitsyn
Faculty of Bioengineering and bioinformatics, Moscow State University
ABSTRACT
For the following minireview different aspects of Ciceribacter thiooxidans’ genome were analysed. These include the number of proteins and RNA’s encoded in the genome, their type and structural features, Chargaff’s second parity rule, GC-count, CDS’s distribution on + and - strand of the DNA molecules, Shine-Dalgarno sequences, stop-codon usage, GC skew and cumulative GC skew of the DNA molecules.
INTRODUCTION
Ciceribacter thiooxidans is a nitrate-reducing thiosulfate oxidizing bacterium. It lives in sulfide-rich anoxic sediment in rivers. Ciceribacter thiooxidans play an important role in nitrogen and sulfur cycling in diverse ecosystems. It belongs to the Rhizobiaceae family most of which strains are nodule-forming symbiotic microorganisms with nitrogen-fixing abilities. Cells are Gram-negative, facultatively chemolithotrophic, facultatively anaerobic, non-spore-forming and rod-shaped (1.0–2.0×0.6–0.8 µm) with a flagellum. Colonies are circular, convex, entire, glistening and semi-translucent, optimal pH is 7.5, nitrate is reduced to the nitrogen gas [1].
MATERIAL AND METHODS
GCF_014126615.1_ASM1412661v1_assembly_stats.txt - file containing different characteristics of the bacteria genome including but not limited to: number of DNA molecules in the cell, GC content, number of scaffolds, length of all DNA molecules present in the cell. It can be found in the supplementary materials (Fig. S1)
GCF_014126615.1_ASM1412661v1_feature_table.txt - file containing information about all the genes and what they encode. There is additional information about their transcripts, such as name and length of the proteins or the type of RNA’s. It can be found in the supplementary materials (Fig. S2). For convenience information from this file is also presented in the Google Sheets file which can be found in the supplementary materials (Fig. S3).
The script written in Python 3.9 was used to calculate the number of each nucleotide, GC skew, 20 nucleotides long sequences before translation initiation starting point and most common 6-mers in them, number of occurrences for each stop-codon. This script can be found in the supplementary materials (Fig. S4)
GCF_014126615.1_ASM1412661v1_genomic.fna - fasta file containing entire DNA sequence of the bacteria. It can be found in the supplementary materials (Fig. S5)
To calculate p-value based on the value of chi-squared test script on the website was used. The link to the website can be found in the supplementary materials (Fig. S6)
GCF_014126615.1_ASM1412661v1_cds_from_genomic.fna.gz - file containing information about all CDS in the genome of Ciceribacter thiooxidans. It can be found in the supplementary materials (Fig. S7)
To assess whether Chargaff’s second parity rule applies to the genome of the Ciceribacter thiooxidans Pearson’s chi-squared test with significance level α = 0.05 was used.
To assess whether protein coding genes are distributed randomly between + and - strands of the genome of the Ciceribacter thiooxidans Pearson’s chi-squared test with significance level α = 0.05 was used.
RESULTS
Basic genome information
The genome contains 2 DNA molecules: 1 chromosome and 1 plasmid. The length of the chromosome is 3661327 nucleotides. The length of the plasmid is 1382059 nucleotides. Both molecules have approximately the same GC-count of 63% and 62% respectively. Genome coverage is 129,0x (Fig. S1).
| Chromosome | Plasmid | |
|---|---|---|
| Length | 3661327 | 1382059 |
| GC-count | 63% | 62% |
Proteome information
There are a total of 4661 proteins encoded in the genome of Ciceribacter thiooxidans. The following histogram made in Google Sheets depicts the number of proteins of different lengths (Histogram 1).
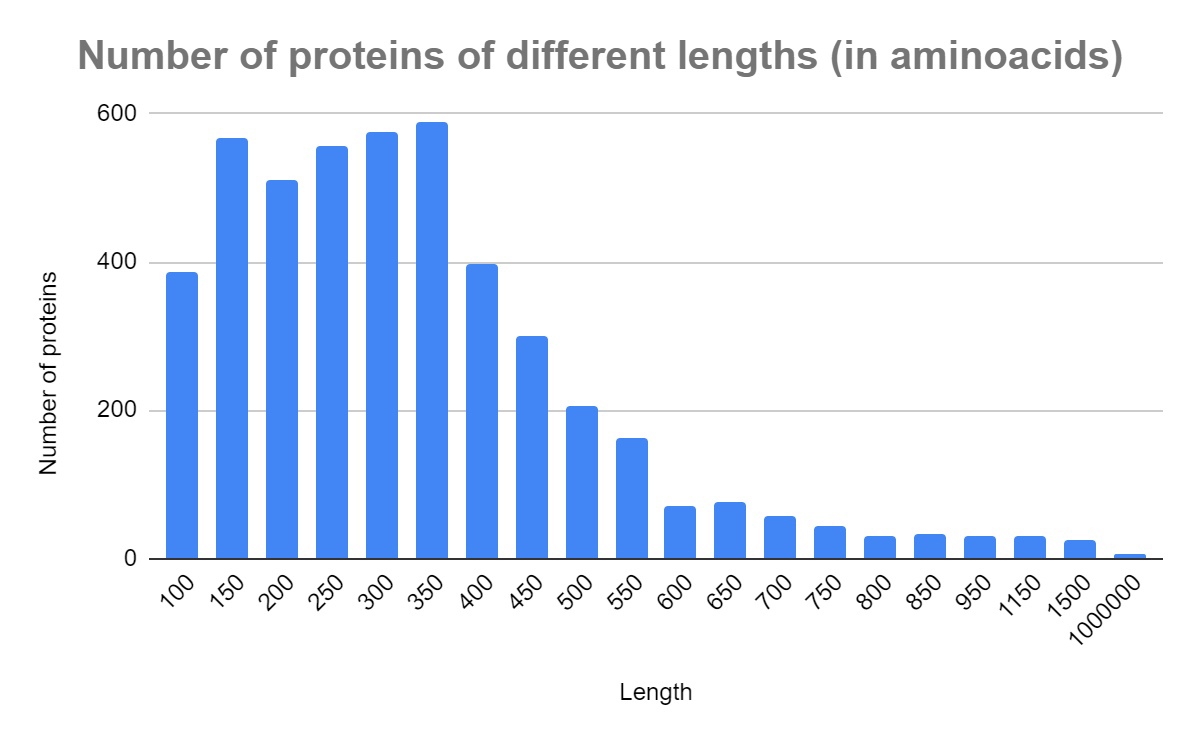
Out of those 4661 proteins 3452 are encoded in the chromosome and 1209 are encoded in the plasmid. There are a total of 2233 proteins encoded on the + strand of the DNA and 2428 encoded on the - strand of the DNA molecule. There are 56 ribosomal, 595 transporter and 491 hypothetical proteins. Transporters make up 12,77% of all proteins and hypothetical proteins make up 10,53% (Fig. S3). More detailed information about the number of different proteins in the chromosome and plasmid can be found in Table 2.
| Chromosome | Plasmid | |
|---|---|---|
| Number of proteins on + strand | 1638 | 595 |
| Number of proteins on - strand | 1814 | 614 |
| Number of transporter proteins | 343 | 252 |
| Number of ribosomal proteins | 56 | 0 |
| Number of hypothetical proteins | 375 | 116 |
| Percentage of transporter proteins | 9.93% | 20.84% |
| Percentage of hypothetical proteins | 10.86% | 9.59% |
RNA’s
There are a total of 68 genes which encode RNA’s. This is expectedly much fewer than the number of protein-encoding genes. All RNA’s are encoded in the chromosome DNA. Information about the types of RNA’s is presented in Table 3.
| RNA genes | Number of these genes |
|---|---|
| All RNA | 68 |
| tRNA | 55 |
| rRNA | 9 |
| tmRNA | 1 |
| RNase_P_RNA | 1 |
| SRP_RNA | 1 |
| 6s/SsrS RNA | 1 |
It should be noted that some tRNA genes have multiple copies- there are 5 tRNA’s-Met with anticodon CAT, 3 tRNA’s-Ala with anticodon CGC, 3 tRNA’s-Ile with anticodon GAT, 2 tRNA’s-Asp with anticodon GTC, 2 tRNA’s-Arg with anticodon TCT, 3 tRNA’s-Ala with anticodon TGC and 2 tRNA’s-Glu with anticodon TTC. This should be accounted for by the researcher who decides to use Ciceribacter thiooxidans as the system for the eukaryotic proteins synthesis (Fig. S3).
Chargaff’s second rule
Chargaff’s second rule states that complementary nucleotides are met with almost equal frequencies in single stranded DNA [2]. Chargaff’s second rule was tested on the chromosome and plasmid of the Ciceribacter thiooxidans using Pearson’s chi-squared test with significance level α = 0.05 [3]. Number of each nucleotide was calculated using a script written in Python (Fig. S4) and FASTA-file containing genome of Ciceribacter thiooxidans (Fig. S5). P-value was calculated using software on the website listed in the supplementary materials (Fig. S6). The results are presented in Table 4
| Chromosome | Plasmid | |
|---|---|---|
| Number of adenines | 675729 | 263164 |
| Number of thymines | 675896 | 261529 |
| Number of cytosines | 1149049 | 430111 |
| Number of guanines | 1160653 | 427255 |
| Chi-squared test value for AT distribution | 0.0206 | 5.0948 |
| Chi-squared test value for GC distribution | 58.3000 | 9.5137 |
| p-value for AT distribution | 0.885874 | 0.023998 |
| p-value for GC distribution | 10 -10 | 0.002039 |
P-value was above chosen significance level only for AT distribution in the chromosome, which means there is no statistical difference between expected and observed number of adenines and thymines in that molecule. The GC and AT distribution in the plasmid are below the significance level, but are still pretty close to it. The most notable difference is in the GC distribution in the chromosome, where p-value is 10 -10.
CDS’s distribution
The hypothesis that there should be the same number of protein-coding genes on + and - strands of the DNA was tested using Pearson’s chi-squared test with the significance level α = 0.05 [3]. The results are presented in Table 5.
| Chromosome | Plasmid | |
|---|---|---|
| Number of CDS’s on + strand | 1638 | 595 |
| Number of CDS’s on - strand | 1814 | 614 |
| Chi-squared test value | 8.9733 | 0.2986 |
| p-value | 0.00274 | 0.584761 |
The statistical difference is only observed in the chromosome, where p-value is 0.00274, meaning CDS’s are not distributed randomly between its + and - strand.
GC-skew
GC skew and cumulative GC skew of both chromosome and plasmid of Ciceribacter thiooxidans were analysed using code written in Python (Fig. S4) and FASTA-file containing genome of Ciceribacter thiooxidans (Fig. S5). The results are presented on the following plots.
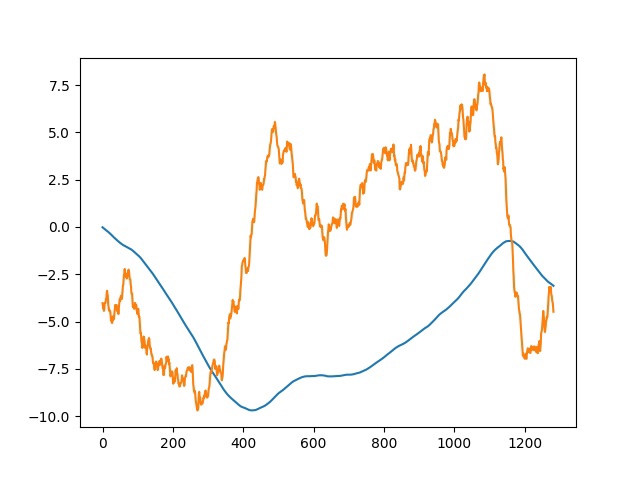
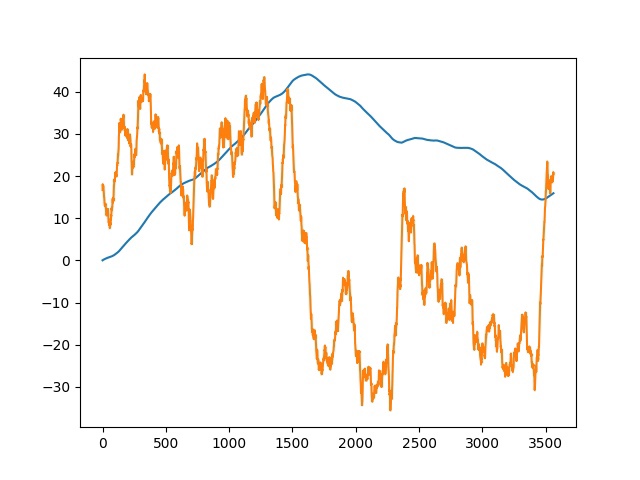
It is known that the maximum value of the cumulative skew corresponds to the terminus, and the minimum value corresponds to the origin of replication [4]. As can be seen from our plots, in the plasmid origin of replication is at around 425000th nucleotide, terminus at around 1158000th nucleotide; in the chromosome origin is at around 1625000th nucleotide, terminus at around 3471000th nucleotide.
6-mers upstream of the translation initiation point
Using script written in Python (Fig. S4) twenty nucleotide long sequences before the translation initiation point were analysed in both chromosome and plasmid DNA of Ciceribacter thiooxidans. Ten of the most frequent 6-mers for each DNA were calculated using protocol written in Python (Fig. S4) and FASTA-file containing genome of Ciceribacter thiooxidans (Fig. S5). The results are presented in Table 6.
| Chromosome | Plasmid | |
|---|---|---|
| 6-mer and number of occurrences |
219 GGAGGA 219 AGGAGA 198 AAGGAG 196 GAGGAG 168 GAAGGA 167 GGAGAA 162 AGGAGG 152 AAAGGA 149 GGGAGG 141 GAAAGG |
133 GGAGGA 111 GAGGAG 101 AGGAGA 97 GGGAGG 74 GGAGAA 70 AGGAGG 70 AAGGAG 64 AGGGAG 61 GAGGAA 60 CGGAGG |
These 6-mers are Shine-Dalgarno sequences [5]. They bind to the 16S rRNA and allow the ribosome to start protein synthesis. As can be seen from Table 5, 19 out of 20 most frequent Shine-Dalgarno sequences consist only of A and G.
Stop codon usage
Number of occurrences for each stop codon was calculated using protocol written in Python (Fig. S5) for both plasmid and chromosome of Ciceribacter thiooxidans. The sequences of CDS’s used for this can be found in supplementary materials (Fig. S7). The results are presented in Table 7.
| Chromosome | Plasmid | |
|---|---|---|
| Number of occurrences of each stop-codon |
TGA 2515 TAA 538 TAG 478 |
TGA 904 TAA 149 TAG 214 |
This is an unexpected result, since in most bacteria TAA stop-codon is the most frequent because it is recognised by both RF1 and RF2 [6]. On the other hand, TGA stop-codon is only recognised by RF2. However, during the translation of mRNA which encodes RF2 ribosomal slippage takes place, so this mRNA also encodes another protein - penicillin-binding protein 1A (Fig. S3). This protein is responsible for penicillin resistance [7]. Ciceribacter thiooxidans was discovered in the sediment of Pearl River Delta in China [1] which is heavily contaminated by different antibiotics [8]. Therefore when Ciceribacter thiooxidans synthesises penicillin-binding protein 1A to protect itself from the high concentration of antibiotics nearby, it also synthesises RF2. It was shown that high concentration of RF2 in the bacteria cell correlates with higher usage of TGA stop-codon [6]. High RF2 concentration caused by water contamination results in much higher TGA stop-codon frequency.
Anticodon usage
Anticodons of tRNA’s present in the Ciceribacter thiooxidans genome were analysed (Fig. S3). The results are presented in Table 8.
| Amino acid | Anticodon of tRNA for this amino acid |
|---|---|
| Arg | CCT, TCT (x2), CCG, ACG, TCG, GCG |
| Leu | CAA, GAG, TAG, TAA, CAG, AAG |
| Ile | GAT (x3), CAT, TAT, AAT |
| Ala | TGC (x3), CGC (x3), GGC, AGC |
| Met | CAT (x4) |
| Val | GAC, TAC, AAC, CAC |
| Lys | CTT, TTT |
| Ser | TGA, GCT, GGA, CGA, AGA, ACT |
| Pro | TGG, GGG, CGG, AGG |
| Glu | TTC (x2), CTC |
| Asn | GTT, ATT |
| Cys | GCA, ACA |
| Trp | CCA |
| Gly | TCC, CCC, GCC, ACC |
| Tyr | GTA, ATA |
| Thr | TGT, GGT, CGT, AGT |
| Asp | GTC (x2), ATC |
| Gln | TTG, CTG |
| His | GTG, ATG |
| Phe | GAA, AAA |
The first observation is that only 41 (CAT anticodon is counted once) out of possible 61 anticodons are present, but obviously all codons encoding amino acids can be translated in the cell. This is achieved because one nucleotide in the anticodon can have more than 1 complementary nucleotide, dictated by Wattson-Crick base pairing rules. This effect is called wobble base pairing, wobble base pairs are presented in Table 9.
| tRNA 5' anticodon base | mRNA 3' codon base (Revised) [9] |
|---|---|
| A | U, C, G, or (A) |
| C | G |
| G | C or U |
| U | A, G, U, or C |
| I | A, C, or U |
| k2C | A |
Another interesting observation is that one tRNA-Ile has CAT anticodon, but tRNA-Ile are only expected to have anticodons GAT, TAT or AAT [10]. CAT anticodon would normally bind to the ATG codon, which encodes methionine. However, the first cytosine in this anticodon is modified by the tRNA lysidine(34) synthetase TilS (Fig. S3), which converts cytosine to lysidine [12]. Lysidine forms base pairs with adenine (Table 9), so the resulting anticodon acts as TAT and can bind to common Ile codons.
SUPPLEMENTARY DATA
S1 GCF_014126615.1_ASM1412661v1_assembly_stats.txt
S2 GCF_014126615.1_ASM1412661v1_feature_table.txt.gz
S4 Python code
S5 GCF_014126615.1_ASM1412661v1_genomic.fna.gz
S6 https://www.socscistatistics.com/pvalues/chidistribution.aspx
S7 GCF_014126615.1_ASM1412661v1_cds_from_genomic.fna.gz
CONFLICT OF INTEREST
The author declares that there are no conflicts of interest.
REFERENCES
- Deng, Tongchu, Xingjuan Chen, Qin Zhang, Yuming Zhong, Jun Guo, Guoping Sun, и MeiyingYR 2017 Xu. «Ciceribacter thiooxidans sp. nov., a novel nitrate-reducing thiosulfate-oxidizing bacterium isolated from sulfide-rich anoxic sediment». International Journal of Systematic and Evolutionary Microbiology 67, issue 11: 4710–15. https://doi.org/10.1099/ijsem.0.002367.
- Fariselli, Piero, Cristian Taccioli, Luca Pagani, и Amos Maritan. «DNA sequence symmetries from randomness: the origin of the Chargaff’s second parity rule». Briefings in Bioinformatics 22, issue 2 (1 of March 2021): 2172–81. https://doi.org/10.1093/bib/bbaa041.
- Pearson, Karl. «X. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling», 1 of July 1900 https://doi.org/10.1080/14786440009463897.
- Grigoriev, Andrei. «Analyzing genomes with cumulative skew diagrams». Nucleic Acids Research 26, vol. 10 (1 of May 1998): 2286–90. https://doi.org/10.1093/nar/26.10.2286.
- «Shine-Dalgarno Sequence - an overview | ScienceDirect Topics». https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/shine-dalgarno-sequence.
- Korkmaz, Gürkan, Mikael Holm, Tobias Wiens, и Suparna Sanyal. «Comprehensive Analysis of Stop Codon Usage in Bacteria and Its Correlation with Release Factor Abundance». The Journal of Biological Chemistry 289, vol. 44 (31 of October 2014): 30334–42. https://doi.org/10.1074/jbc.M114.606632.
- Sousa Oliveira, K. de, L. A. de Lima, N. B. Cobacho, S. C. Dias, и O. L. Franco. «Chapter 2 - Mechanisms of Antibacterial Resistance: Shedding Some Light on These Obscure Processes?» В Antibiotic Resistance, red. Kateryna Kon и Mahendra Rai, 19–35. Academic Press, 2016. https://doi.org/10.1016/B978-0-12-803642-6.00002-2.
- Xu, Weihai, Wen Yan, Xiangdong Li, Yongde Zou, Xiaoxiang Chen, Weixia Huang, Li Miao, Ruijie Zhang, Gan Zhang, и Shichun Zou. «Antibiotics in Riverine Runoff of the Pearl River Delta and Pearl River Estuary, China: Concentrations, Mass Loading and Ecological Risks». Environmental Pollution (Barking, Essex: 1987) 182 (November 2013): 402–7. https://doi.org/10.1016/j.envpol.2013.08.004.
- Murphy IV, Frank V; Ramakrishnan, V (21 November 2004). "Structure of a purine-purine wobble base pair in the decoding center of the ribosome". Nature Structural & Molecular Biology. 11 (12): 1251–1252. doi:10.1038/nsmb866. PMID 15558050. S2CID 27022506.
- «Amino Acid Translation Table», 29 of May 2020 https://web.archive.org/web/20200529000711/http://sites.science.oregonstate.edu/genbio/otherresources/aminoacidtranslation.htm.
- Suzuki, Tsutomu, и Kenjyo Miyauchi. «Discovery and Characterization of TRNAIle Lysidine Synthetase (TilS)». FEBS Letters 584, vol. 2 (21 of January 2010): 272–77. https://doi.org/10.1016/j.febslet.2009.11.085.