Mini-review
Genome and proteome analysis of the Gram-negative bacterium Coxiella burnetii RSA 493
Elisaveta A. Nikishina
Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, Moscow, Russia
Abstract: We have analysed the genome and the proteome of the Gram-negative bacterium Coxiella burnetii RSA 493
Motivation: conduct research using various methods
Results: information about the bacterium has been clarified
Contact: lnik04@fbb.msu.ru
1. Introduction
Coxiella burnetii is a Gram-negative bacterium which belongs to the class gamma-proteobacteria, genus Coxiella, species Coxiella burnetii. (Fig. 1) It was found in Montana, USA, 1935. Cells of Coxiella burnetii have Nine Mile strains – the virulent phase I (RSA 493) and avirulent phase II (RSA 439) [1]. The extracellular matrix of this bacterium is almost the same as that of other gram-negative bacteria. There are antigenic and compositional differences between the virulent phase I and the avirulent phase II. The virulent phase is highly contagious, but phase II – not, it occurs only in laboratory strains.
Q fever disease of humans and other animals is caused by this gamma-proteobacteria. In humans,Q fever can manifest itself in both acute and chronic forms. It usually presents like the flu but it can progress to pneumonia [2]. Coxiella burnetii is resistant to environmental factors such as desiccation, heat and disinfection, so it has a very long life [1].
2. Methods
2.1 Genome analysis
2.1.1 The number and names of the DNA that make up the genome and GC-content
Information about DNA length and GC-content was taken from the NCBI database from file GCF_000007765.2_ASM776v2_assembly_stats.txt (Supplementary materials S1).
2.2 Proteome analysis
Proteome analysis was made in Google tables by sorting information. The table of genome features of Coxiella burnetii RSA 493 was imported into Google Sheets on the sheet “GCF_000007765.2_ASM776v2_feature_table.txt” from NCBI database (Supplementary materials S2). We found information by using keywords (“with_protein” and “protein_coding”) in column B. To find information about direct and complementary chains, we sorted information in column J (counted “+” for direct chain and “-” for complementary).
2.3 RNA genes analysis
RNA genes analysis was made in Google tables by sorting information. The table of genome features of Coxiella burnetii RSA 493 was imported into Google Sheets on the sheet “GCF_000007765.2_ASM776v2_feature_table.txt” from NCBI database. Genes were found with a combination of keywords in columns A and B (“gene” and “RNA”)(Supplementary materials S4).
2.4 Analysis of protein-coding nucleotide sequences
Information about the sequence was taken from the NCBI database from file ‘GCF_000007765.2_ASM776v2_genomic.fna’. (Supplementary materials S5). Protein-coding nucleotide sequences analysis was made by using my python program (Supplementary materials S6). It worked with the file mentioned above and counted the usage of every codon and printed it.
3. Results
3.1 Genome analysis
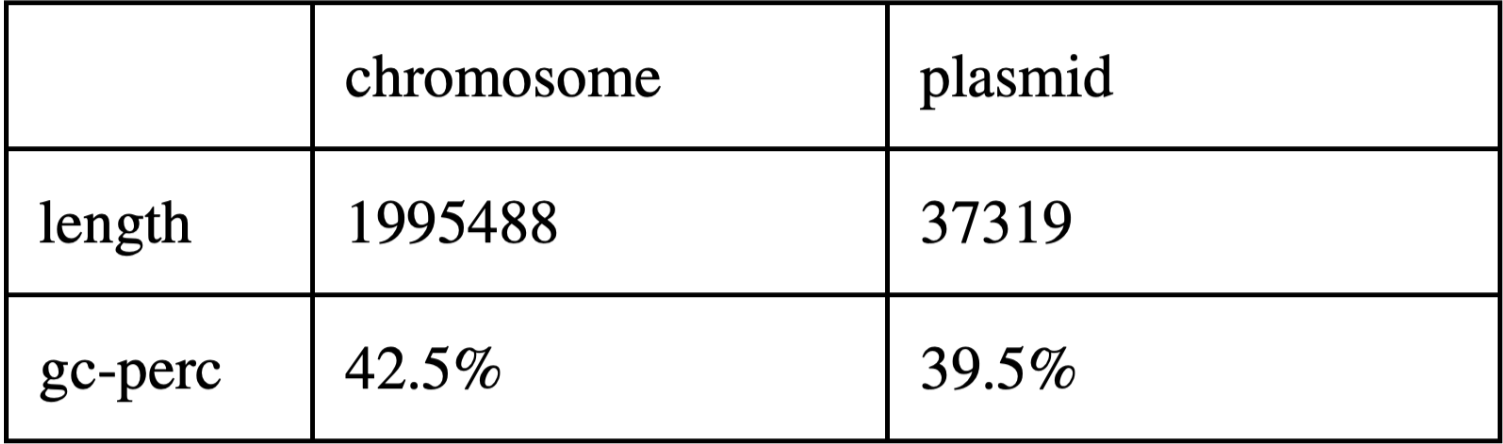
Genome of Coxiella burnetii RSA 493 consists of 1 chromosome and 1 plasmid.
3.2 Proteome analysis
3.2.1 Protein length
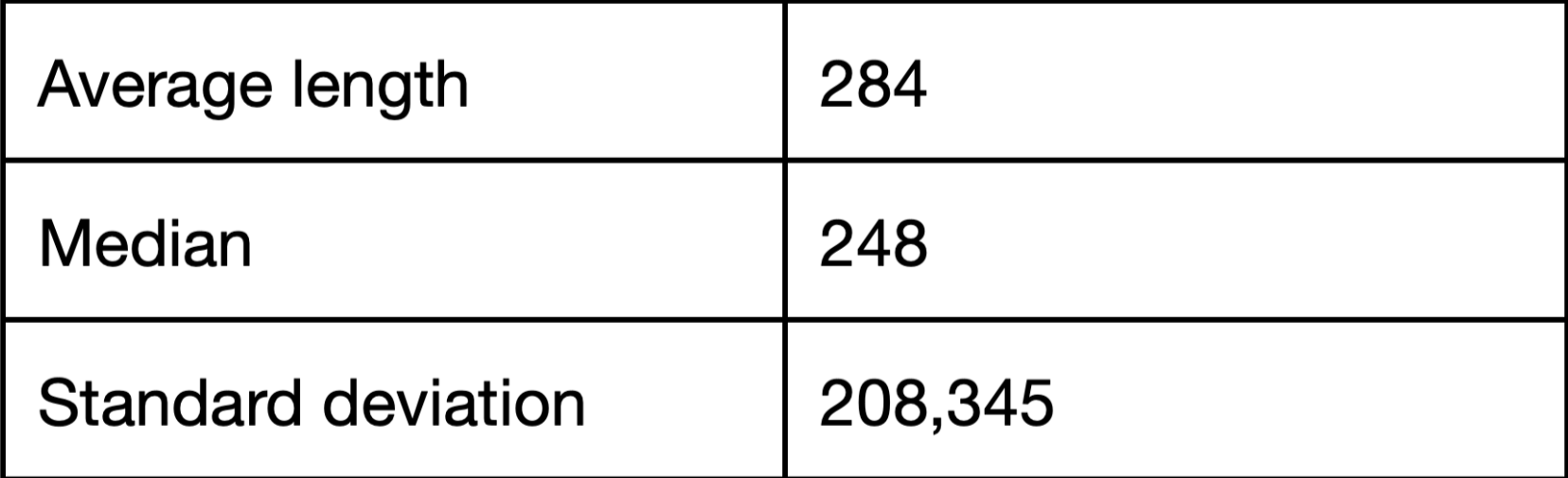
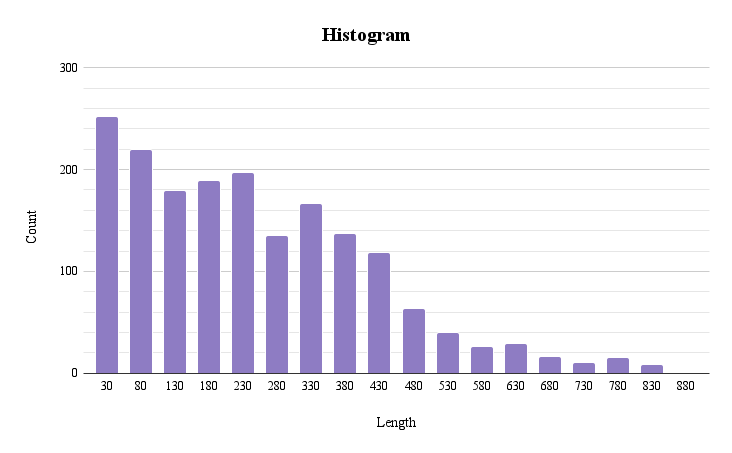
This distribution histogram of protein length was made with the help of Google tables (Fig 2). Average length is 284 amino acids, median is 248 amino acids. Standard deviation - 208,345. (Supplementary materials S3)
This histogram shows the distribution of protein lengths. In each column, the proteins are distributed in increments of 50. The first represents the number of proteins with a length from 0 to 30, the second with a length from 30 to 80, and so on.
3.2.2 The number of protein genes encoded on the direct and complementary chain, ribosomal proteins, hypothetical proteins and transport proteins
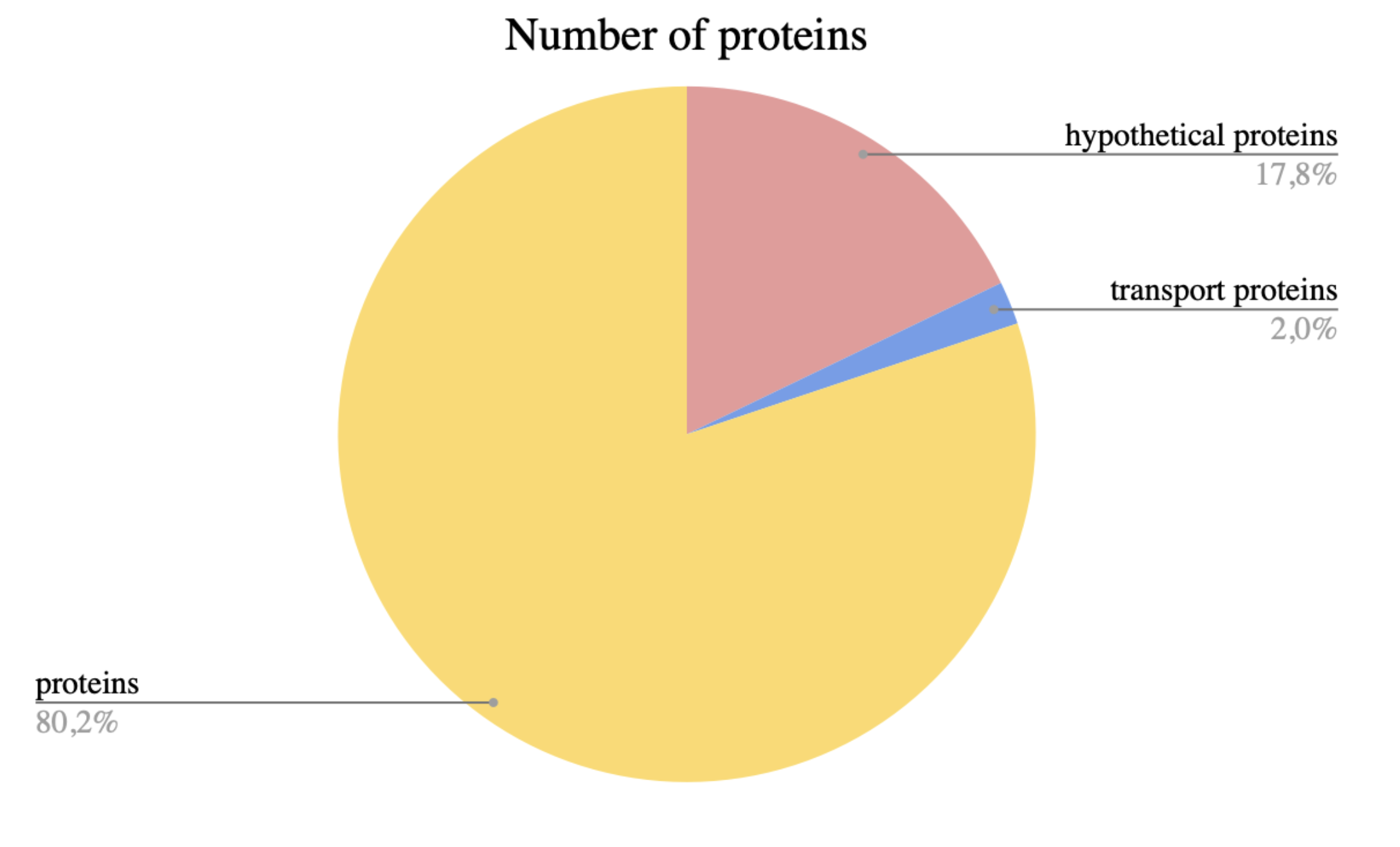
The number of genes encoded on the direct and complementary chain was compared on google tables using special sorting. As a result, it turned out that on the direct chain there are 1938 protein genes, on complementary - 1728 genes. The number of ribosomal proteins was 66. There were 654 hypothetical proteins and this was 17.84% of the total number. While there were 74 transport proteins and this was only 2% of the total number of proteins (Supplementary materials S4).
3.3 RNA genes analysis
The number of RNA genes is much smaller than the number of protein genes. There are 66 of them; this is 0,764% of the total number of protein genes.Moreover, in the ribosome of Coxiella burnetii RSA 493 there are 12 rRNA. The number of tRNA is 84.
3.4 Analysis of protein-coding nucleotide sequences
The result of my python code is the printed number of the usage of every protein- coding nucleotide. Number of uses of “TAA”, “TAG”, “TGA” is 0 because they are stop-codons.
Result:
AAA 28611, AAC 11740, AAG 13231, AAT 20599, ACA 8040, ACC 8072, ACG 8534, ACT 6993, AGA 8500, AGC 10364, AGG 7323, AGT 6942, ATA 12224, ATC 12150, ATG 10443, ATT 20984, CAA 14394, CAC 7094, CAG 6934, CAT 10686, CCA 10012, CCC 7505, CCG 8717, CCT 7321, CGA 10083, CGC 11005, CGG 8572, CGT 8480, CTA 6559, CTC 6797, CTG 7054, CTT 13125, GAA 14435, GAC 6026, GAG 6729, GAT 12102, GCA 9680, GCC 9377, GCG 10930, GCT 10290, GGA 8778, GGC 9311, GGG 7481, GGT 8022, GTA 6640, GTC 6068, GTG 7008, GTT 11584, TAA 0, TAC 6716, TAG 0, TAT 12601, TCA 11673, TCC 8363, TCG 10244, TCT 8515, TGA 0, TGC 9722, TGG 10119, TGT 8063, TTA 17157, TTC 13868, TTG 14483, TTT 27734
Acknowledgements
We express our gratitude to colleagues from the faculty of Bioengineering and Bioinformatics in MSU.
Supplementary materials
S1. NCBI file with results of Genome analysis of Coxiella burnetii RSA 493
S2. Table with Genome features which was taken as a base for analysis
Genome features of Coxiella burnetii RSA 493
S3. Table with sorted information about length of proteins and histogram
Table with results of Proteome analysis of Coxiella burnetii RSA 493
S4.Table with sorted information about proteins and RNA genes which was necessary for analysis
Table with results of proteome analysis
S5. File with sequences which was used for doing analysis
GCF_000007765.2_ASM776v2_genomic.fna.gz
S6.Code which was used for doing analysis of protein-coding nucleotide sequences
Python code for analysis of protein-coding nucleotide sequences
References
[1] Ludovit Skultetya, Martin Hajduch, Gabriela Flores-Ramirez, Ján A. Miernykc, Fedor Ciampor, Rudolf Toman, Zuzana Sekeyova. Proteomic comparison of virulent phase I and avirulent phase II of Coxiella burnetii, the causative agent of Q fever. 1874-3919/$ – see front matter, 2011 Elsevier B.V. doi: 10.1016/j.jprot.2011.05.017
[2] K. E. Russell-Lodrigue, G. Q. Zhang, D. N. McMurray, and J. E. Samuel. Clinical and Pathologic Changes in a Guinea Pig Aerosol Challenge Model of Acute Q Fever. INFECTION AND IMMUNITY, Nov. 2006, p. 6085–6091 Vol. 74, No. 11 0019-9567/06/$08.00+0 doi:10.1128/IAI.00763-06 Copyright 2006, American Society for Microbiology.