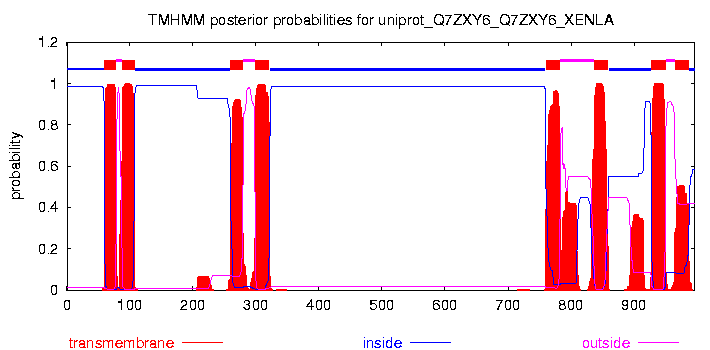
# uniprot_Q7ZXY6_Q7ZXY6_XENLA Length: 996 # uniprot_Q7ZXY6_Q7ZXY6_XENLA Number of predicted TMHs: 8 # uniprot_Q7ZXY6_Q7ZXY6_XENLA Exp number of AAs in TMHs: 177.60819 # uniprot_Q7ZXY6_Q7ZXY6_XENLA Exp number, first 60 AAs: 0.86623 # uniprot_Q7ZXY6_Q7ZXY6_XENLA Total prob of N-in: 0.98658 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 inside 1 59 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 60 78 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 outside 79 87 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 88 107 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 inside 108 259 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 260 279 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 outside 280 298 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 299 321 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 inside 322 759 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 760 782 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 outside 783 836 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 837 859 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 inside 860 927 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 928 950 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 outside 951 964 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 TMhelix 965 987 uniprot_Q7ZXY6_Q7ZXY6_XENLA TMHMM2.0 inside 988 996

# plot in postscript,
script for making the plot in gnuplot,
data for plot