|
|
|
Результаты упражнений
Моделирование белка в комплексе с лигандом по гомологии
Выравнивание построенное Clastal и модифицированное для моделирование программой modeller
>P1;seq
sequence:РҐРҐРҐРҐРҐ::::::: 0.00: 0.00
KVYERCELAAAMKRLGLDNYRGYSLGNWVCAAKYESNFNTQATNRNTDGS
TDYGILEINSRWWCNDGKTPGAKNVCGIPCSVLLRSDITEAVKCAKRIVS
DGNGMNAWVAWRNRCKGTDVSRWIRGCRL/.*
>P1;1lmp
structureX:1LMP.pdb:1 :A: 130 :A:undefined:undefined:-1.00:-1.00
KVYDRCELARALKASGMDGYAGNSLPNWVCLSKWESSYNTQATNRNTDGS
TDYGIFQINSRYWCDDGRTPGAKNVCGIRCSQLLTDDLTVAIRCAKRVVL
DPNGIGAWVAWRLHCQNQDLRSYVAGCGV/.*
Из файла 1lmp.pdb были удалены вся вода и CONTACTS
А лиганд изменен следующим образом.
HETATM 1001 C1 NAG A 130 40.573 45.941 30.725 1.00 16.35 C
HETATM 1002 C2 NAG A 130 40.599 47.461 31.105 1.00 49.86 C
HETATM 1003 C3 NAG A 130 39.697 47.780 32.309 1.00 29.37 C
HETATM 1004 C4 NAG A 130 38.249 47.104 32.228 1.00 20.77 C
HETATM 1005 C5 NAG A 130 38.436 45.626 31.880 1.00 22.06 C
HETATM 1006 C6 NAG A 130 37.186 44.958 31.539 1.00 18.57 C
HETATM 1007 C7 NAG A 130 42.034 48.701 30.178 1.00 25.62 C
HETATM 1008 C8 NAG A 130 43.346 49.262 30.759 1.00 42.60 C
HETATM 1009 N2 NAG A 130 41.695 47.858 31.171 1.00 42.79 N
HETATM 1010 O3 NAG A 130 39.266 49.181 32.311 1.00 46.30 O
HETATM 1011 O4 NAG A 130 37.469 47.416 33.391 1.00 26.43 O
HETATM 1012 O5 NAG A 130 39.246 45.423 30.710 1.00 20.34 O
HETATM 1013 O6 NAG A 130 36.383 45.653 30.651 1.00 21.58 O
HETATM 1014 O7 NAG A 130 41.594 49.122 29.081 1.00 58.35 O
HETATM 1015 C1A NAG A 130 43.023 42.938 27.078 1.00 24.56 C
HETATM 1016 C2A NAG A 130 41.492 42.604 27.308 1.00 30.33 C
HETATM 1017 C3A NAG A 130 40.822 43.798 27.982 1.00 13.79 C
HETATM 1018 C4A NAG A 130 41.639 44.440 29.207 1.00 16.75 C
HETATM 1019 C5A NAG A 130 43.116 44.565 28.850 1.00 32.65 C
HETATM 1020 C6A NAG A 130 43.971 44.853 30.023 1.00 38.78 C
HETATM 1021 C7A NAG A 130 40.423 40.786 26.468 1.00 12.52 C
HETATM 1022 C8A NAG A 130 39.899 40.642 25.035 1.00 12.39 C
HETATM 1023 N2A NAG A 130 40.892 42.050 26.534 1.00 17.85 N
HETATM 1024 O3A NAG A 130 39.563 43.515 28.603 1.00 15.77 O
HETATM 1025 O4A NAG A 130 41.024 45.731 29.426 1.00 17.85 O
HETATM 1026 O5A NAG A 130 43.596 43.274 28.397 1.00 20.51 O
HETATM 1027 O6A NAG A 130 43.753 44.063 31.167 1.00 56.49 O
HETATM 1028 O7A NAG A 130 40.336 39.746 27.152 1.00 16.98 O
HETATM 1029 C1B NAG A 130 46.881 40.478 24.981 1.00 29.15 C
HETATM 1030 C2B NAG A 130 46.805 41.995 25.238 1.00 28.47 C
HETATM 1031 C3B NAG A 130 45.696 42.310 26.245 1.00 33.80 C
HETATM 1032 C4B NAG A 130 44.322 41.736 25.871 1.00 33.06 C
HETATM 1033 C5B NAG A 130 44.299 40.337 25.216 1.00 33.05 C
HETATM 1034 C6B NAG A 130 43.345 40.460 24.056 1.00 28.22 C
HETATM 1035 C7B NAG A 130 48.758 43.117 26.079 1.00 34.25 C
HETATM 1036 C8B NAG A 130 48.543 44.254 25.032 1.00 34.25 C
HETATM 1037 OB NAG A 130 45.558 39.988 24.562 1.00 34.25 O
HETATM 1038 O3B NAG A 130 45.624 43.701 26.666 1.00 32.94 O
HETATM 1039 O4B NAG A 130 43.524 41.622 27.086 1.00 26.50 O
HETATM 1040 O6B NAG A 130 43.599 41.602 23.184 1.00 25.99 O
HETATM 1041 O7B NAG A 130 49.919 43.191 26.673 1.00 34.25 O
HETATM 1042 N2B NAG A 130 47.855 42.106 26.370 1.00 34.25 N
HETATM 1043 O1L NAG A 130 47.236 40.018 26.007 1.00 20.00 O
Управляющий скрипт
from modeller.automodel import *
class mymodel(automodel):
def special_restraints(self, aln):
rsr = self.restraints
for ids in (('O:107:A', 'N2A:130:B'),
('N:109:A', 'O6B:130:B'),
('NE1:62:A','O6A:130:B')):
atoms = [self.atoms[i] for i in ids]
rsr.add(forms.upper_bound(group=physical.upper_distance,
feature=features.distance(*atoms), mean=3.5, stdev=0.1))
env = environ()
env.io.hetatm = True
a = mymodel(env, alnfile='aln.ali', knowns=('1lmp'), sequence='seq')
a.starting_model = 1
a.ending_model = 5
a.make()
Было получено 5 моделей, которые были оценены критерием WHATIF.
Structure Z-scores, positive is better than average:
Ramachandran plot appearance : 0.606 0.929 -0.051 0.207 0.920
chi-1/chi-2 rotamer normality : -2.363 -2.071 -2.861 -2.495 -3.045
Backbone conformation : -1.493 -1.600 -1.428 -1.287 -1.544
RMS Z-scores, should be close to 1.0:
Side chain planarity : 0.715 0.586 0.322 0.469 0.743
Improper dihedral distribution : 1.385 1.199 1.144 0.926 0.933
На мой взгляд лучшая первая модель.
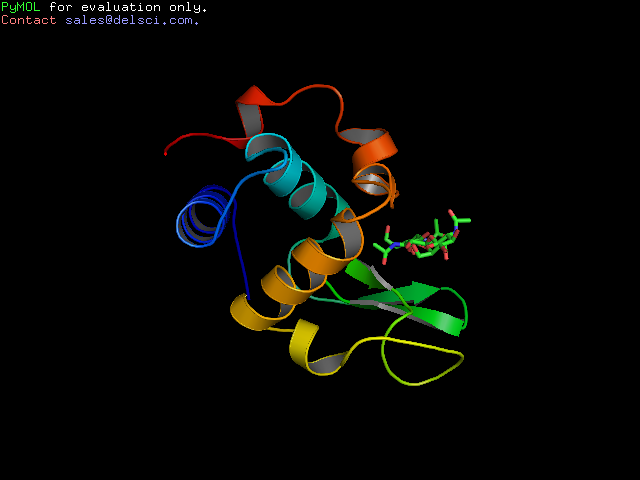
|

