PR10
1. Characteristics of the two best alignments of the polyprotein poliovirus and the foot and mouth disease virus polyprotein
| OC1 | OC2 | ID1 | AC1 | ID2 | AC2 | Score | % Identity | % Positive | Gaps (%) | Site length1 | Site length2 |
| poliovirus | aphthovirus | POLG_POL2L | P06210 | POLG_FMDVT | P15072 | 253 bits(645) | 29 | 48 | 8 | 614 | 643 |
| poliovirus | aphthovirus | POLG_POL2L | P06210 | POLG_FMDVT | P15072 | 158 bits(400) | 30 | 44 | 15 | 409 | 430 |
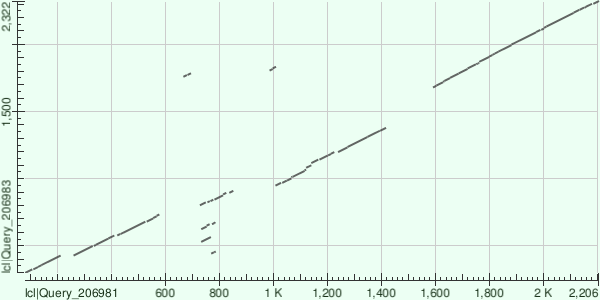
2. Map of local similarity of the polyprotein poliovirus and the foot and mouth disease virus polyprotein
3. Comparison of score alignment with random in homologous and non-homologous proteins
| H/NH | ID1 | ID2 | Score | Median | Q1 | Score (B) | p |
| homologous | GLGB_ECOLI | GLGB_SYNE7 | 92.5 | 92.5 | 99.75 | 1 | 0.5 |
| nonhomologous | PRS10_ECOLI | GLGB_SYNE7 | 24.5 | 31 | 34.75 | 2.63 | 2^(-2.73) |
4. Characterization of alignment of ANS42615.1 homologues from SW bank (BLAST)
| ID | AC | Organism | Score | % Identity | % Positive | Gaps (%) | Site length | Expect | %Coverage |
| YFEA_YERPE | Q56952 | Yersinia pestis | 491 bits(1265) | 80 | 86 | 2 | 295 | 6×10^(-176) | 94 |
| Y362_HAEIN | Q57449 | Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) | 442 bits(1138) | 74 | 83 | 0 | 286 | 7×10^(-157) | 91 |
© Grigoreva Masha