Реконструкция филогении
для реконструкции филогенетического дерева бы выбран белок 16S рРНК-метилтрансфераза H с мнемоникой RSMH
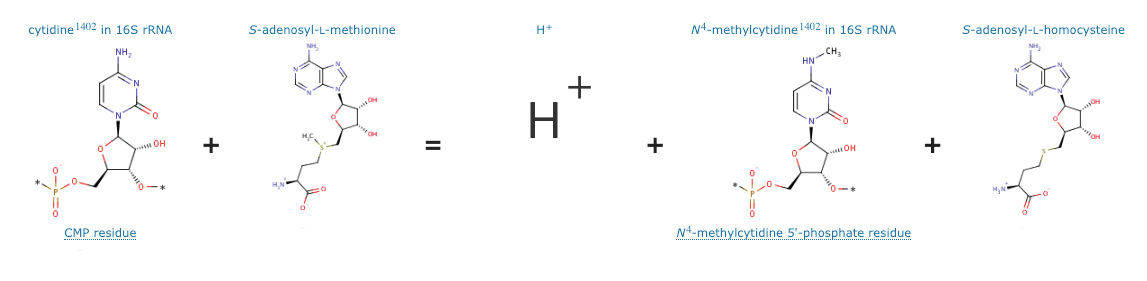
Fig1. Catalytic activity
список идентификаторов белков:
| Индикатор | Название белка | Кодирующий ген | Штамм |
| RSMH_PROMH | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, PMI2078 | Proteus mirabilis (strain HI4320) |
| RSMH_SACD2 | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, Sde_0840 | Saccharophagus degradans (strain 2-40 / ATCC 43961 / DSM 17024) |
| RSMH_ACICJ | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, Acry_0055 | Acidiphilium cryptum (strain JF-5) |
| RSMH_PARDP | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, Pden_0583 | Paracoccus denitrificans (strain Pd 1222) |
| RSMH_ROSDO | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, RD1_3368 | Roseobacter denitrificans (strain ATCC 33942 / OCh 114) (Erythrobacter sp. (strain OCh 114)) (Roseobacter denitrificans) |
| RSMH_BARHE | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, BH11320 | Bartonella henselae (strain ATCC 49882 / DSM 28221 / Houston 1) (Rochalimaea henselae) |
| RSMH_AGRFC | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, Atu2102, AGR_C_3815 | Agrobacterium fabrum (strain C58 / ATCC 33970) (Agrobacterium tumefaciens (strain C58)) |
| RSMH_RHIME | Ribosomal RNA small subunit methyltransferase H | rsmH mraW, R02184, SMc01858 | Rhizobium meliloti (strain 1021) (Ensifer meliloti) (Sinorhizobium meliloti) |
Последовательности этих белковбыли выровнены с помощью программы:
muscle -in term4pr2-notmus.fasta -out term4pr2-mus.fasta
На всякий случай привожу данные вывода этой программы в командную строку:
| term4pr2-notmus 8 seqs, max length 346, avg length 327 |
| 00:00:00 22 MB(-3%) Iter 1 100.00% K-mer dist pass 1 |
| 00:00:00 22 MB(-3%) Iter 1 100.00% K-mer dist pass 2 |
| 00:00:01 25 MB(-4%) Iter 1 100.00% Align node |
| 00:00:01 25 MB(-4%) Iter 1 100.00% Root alignment |
| 00:00:01 26 MB(-4%) Iter 2 100.00% Refine tree |
| 00:00:01 26 MB(-4%) Iter 2 100.00% Root alignment |
| 00:00:01 26 MB(-4%) Iter 2 100.00% Root alignment |
| 00:00:01 26 MB(-4%) Iter 3 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 4 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 5 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 5 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 6 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 7 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 8 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 9 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 9 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 10 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 11 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 12 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 13 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 14 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 15 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 16 100.00% Refine biparts |
| 00:00:01 26 MB(-4%) Iter 17 100.00% Refine biparts |
Далее я открыла выравнивание в MEGA (методом "Analyze") и реконструируировала филогению, предварительно удалив все лишнее из названий последовательностей, в том числе и мнемонику белков, оставив только мнемонику видов (файл term4pr2-mus-new.fasta)
Затем я провела построение с помощью 5ти различных алгоритмов.
UPGMA
Это метод:
+ прямой
+ использует молекулярные часы (строит укорененное ультраметрическое дерево)
+ дистанционный
+ реконструирует длины ветвей.
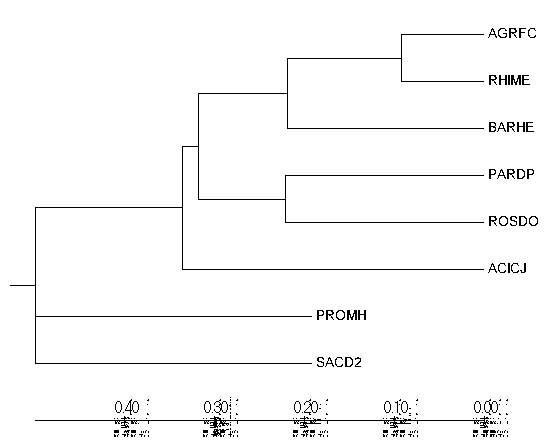 | 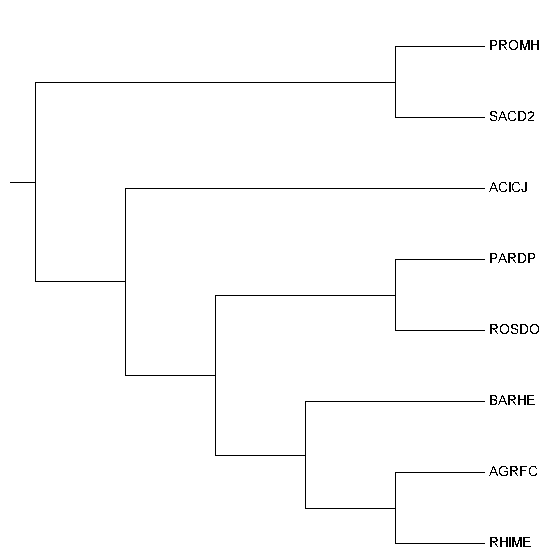 |
| Fig2. Tree by UPGMA | Original Tree from pr1 |
Neighbor-Joining
Это метод:
+ прямой
+ не использует молекулярные часы (строит неукорененное дерево)
+ дистанционный
+ реконструирует длины ветвей.
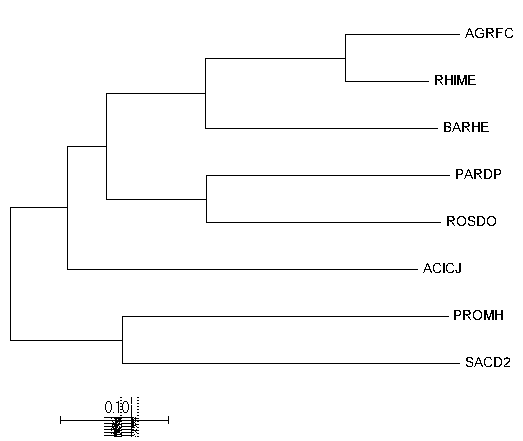 | 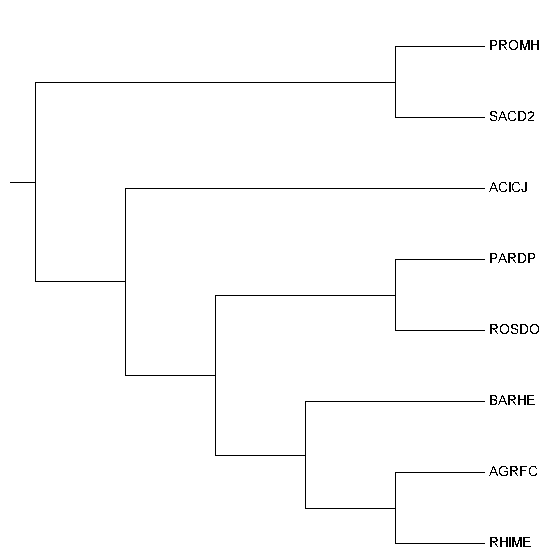 |
| Fig3. Tree by Neighbor-Joining | Original Tree from pr1 |
Минимальной эволюции
Это метод:
+ переборный
+ может использовать молекулярные часы
+ дистанционный
+ реконструирует длины ветвей.
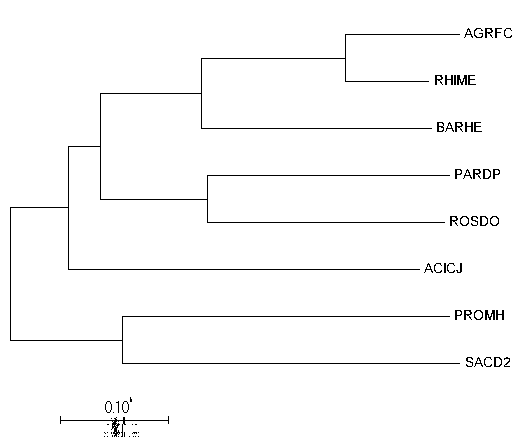 | 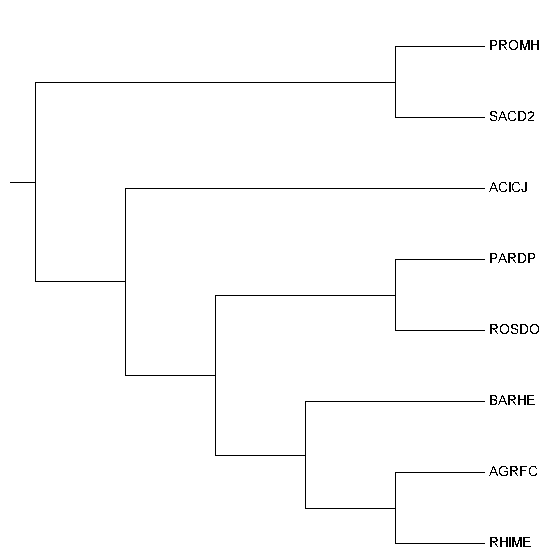 |
| Fig4. Tree by Minimum-Evolution | Original Tree from pr1 |
Максимальной экономии
Это метод:
+ переборный
+ не может использовать молекулярные часы
+ символьный
+ не реконструирует длины ветвей.
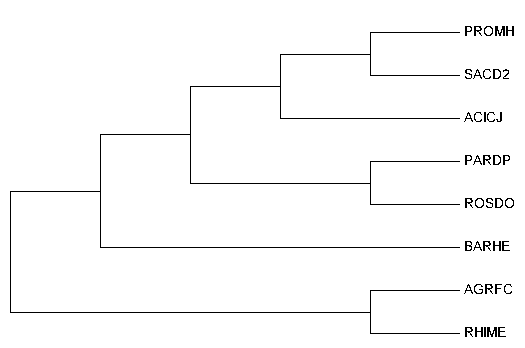 | 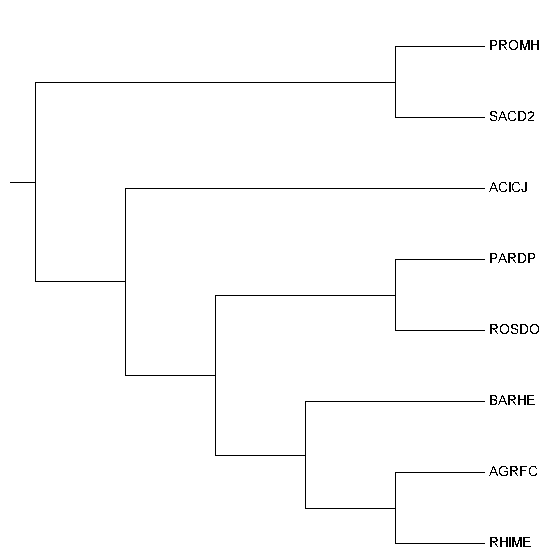 |
| Fig4. Tree by Maximum-Parsimony | Original Tree from pr1 |
Наибольшего правдоподобия
Это метод:
+ переборный
+ может использовать молекулярные часы
+ символьный
+ реконструирует длины ветвей.
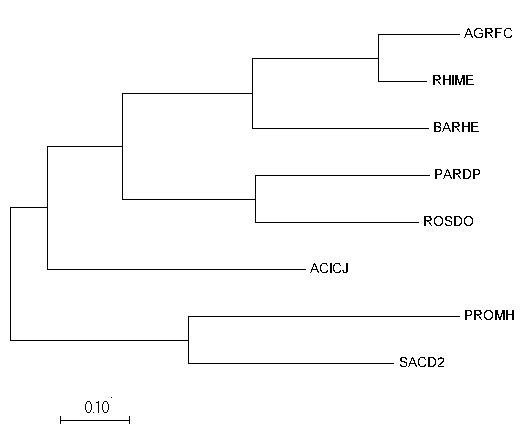 | 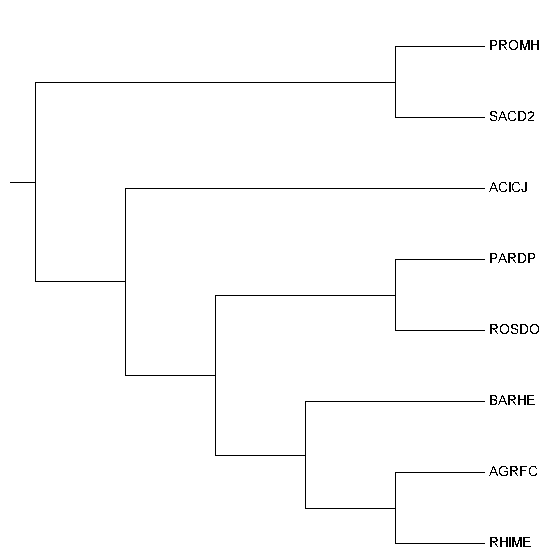 |
| Fig4. Tree by Minimum-Likelihood | Original Tree from pr1 |
© Grigorjeva Masha