Analysis of Oscillatoria acuminata PCC 6304 genome and proteome
Loev M.
Deparment of Bioengeneering and Bioinformatics, Moscow State University
Abstract
In that brief review of Oscillatoria acuminata PCC 6304 genome and proteome I’ve mainly estimated the distribution of different characteristics like protein length, start- and stop-codon usage and GC-content, also to find the possible connection between stop-codon and level of protein expression.
Keywords: Oscillatoria, stop-codon, GC-content, rRNA clusters
Introduction
Oscillatoria acuminata PCC 6304 is a photosynthetic filamentous cyanobacterium living in the fresh water. Genus and species names stem from the oscillation in its movement and pointed end of the filament. Object is of interest because it has a photosynthetic apparatus and produces the butylated hydroxytoluene (BHT), which is used as an antioxidant in chemical production and food industry (E321). The genome sequence is collected from chromosome and two plasmids containing, in total, 7 901 825 bp. with 5851 encoded proteins. Length of the latter differs from 28 to 3704 bp. Current review was conducted with Google Sheets, bash and Python tools. The review was made to put into practice all what was learnt during the first term and try to find some interesting dependencies or reasons of what previously was perceived as obvious. Mainly it gives a cursory glance, but later, I hope, it will be supplemented[1][2][3][4]
Methods
Review is based on two main files: FASTA file with whole genome, collected from one chromosome and two plasmids(pOSCIL6304.01 and pOSCIL6304.02), and a feature table with CDS with protein. They were collected from NCBI database [4] Excel was mainly used for visuals and sorting the outcome of Python programmes. Histogram of protein length was made with the data from the feature table. I’ve made intervals in steps of 45 amino-acids and counted the number of proteins which length fits that gap. Python was used to make CDS parts of genomes from FASTA files and tables. It was important to take the strand into account, because it would end up with completely different results (f.e. I’d get some strange start- or stop-codons). I’ve collected the data of different start and stop-codon frequencies, and then the names of proteins they delimit. This was made to find the possible dependencies between the start(stop)-codon and role of protein. Then I made a file with the areas between CDS, so I can analyze the GC-content in different parts of the genome to find whether they correlate or not. In the Internet I found the information about proteins (UniProt) and some articles with the information needed to understand the outcome. Also I used a website that compares two texts to find the differences between the rRNA coding sequences.[17] All Python programmes used for the review can be seen in the Google Colaboratory [18]
Results
1.Proteome
1.1 rRNA-clasters
DNA contains information not only about proteins, but about a wider range of matter. One of them is rRNA. rRNA is a ribosomal RNA, it is a component of ribosomes and it takes part in protein synthesis. [16] Gene coding rRNA and placed close to each other for so called clusters. rRNA clasters are also called rDNA [16].Result of the analysis of Osclillatoria acuminata PCC 6304 genome is shown in Table 1. As it can be seen, there are three clusters, each made up of 3 types of rRNA (5S, 16S, 23S). Two of them are placed on the positive strand and one on the last, all are located in the chromosome.
| rRNA type | Length between parts of claster in bp | Length in bp | |
|---|---|---|---|
| Claster 1 (+) | 16S | - | 1490 | 23S | 345 | 2889 |
| 5S | 63 | 118 | |
| Claster 2 (+) | 16S | - | 1490 |
| 23S | 550 | 2888 | |
| 5S | 72 | 118 | |
| Claster 3 (-) | 5S | 63 | 118 |
| 23S | 345 | 2888 | |
| 16S | - | 1491 |
Interesting, that all the clusters have one sequence of rRNA types (claster 3 should be investigated in the opposite direction). Another interesting feature is that clasters 1 and 3 have equal relative placing of their parts. But they still have some difference in the parts length. How similar are their nucleotide sequences? It turns out that they differ in 16S and 23S with, in total, 37 different parts. 5S in all clusters is surprisingly equal.
1.2 Protein length distribution
Proteome of Oscillatoria acuminata PCC 6304 includes 5851 proteins.
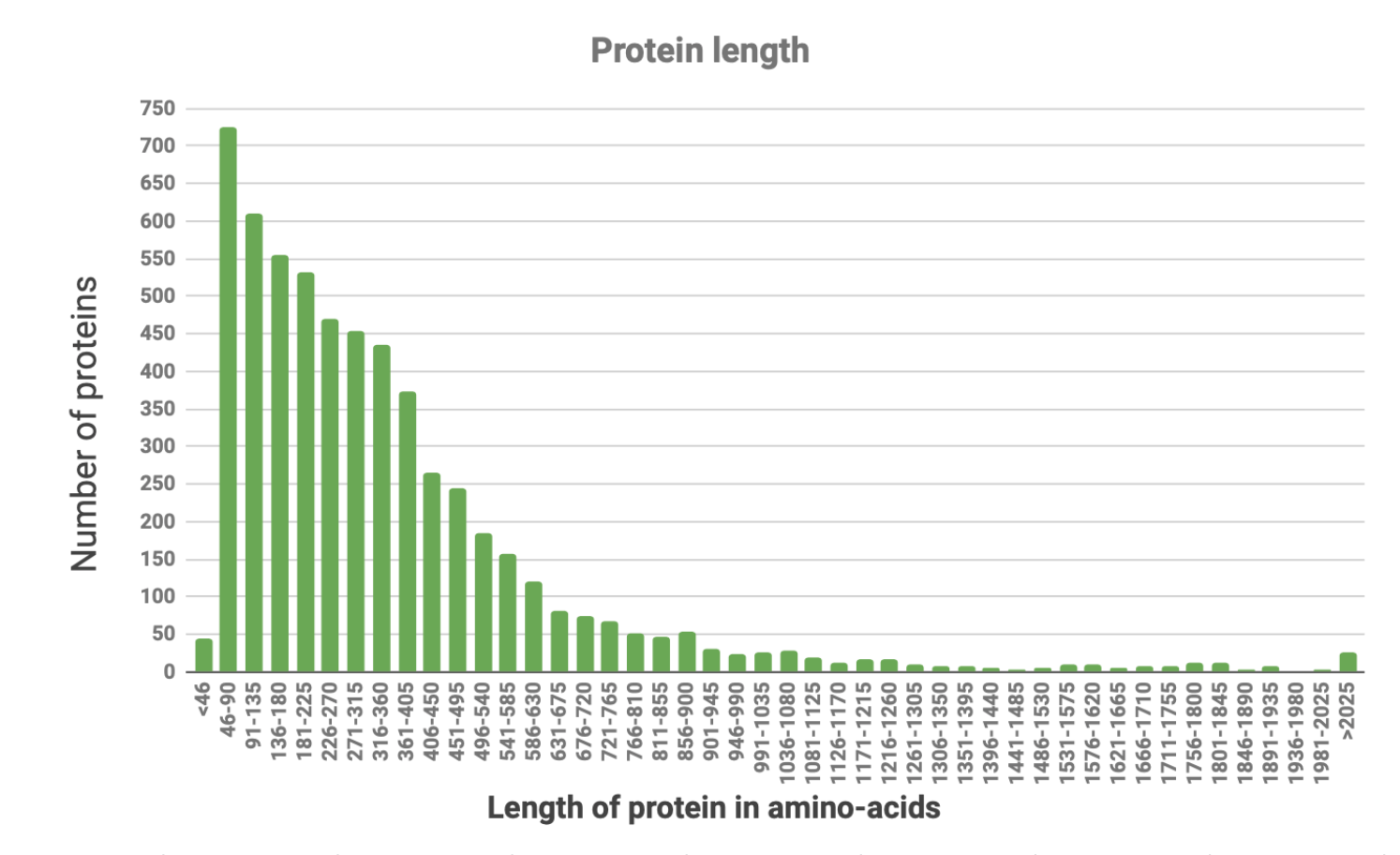
Picture 1 shows that the major part (57,17%) of all proteins are made up of 46-315 amino-acids.
1.3 The longest, the shortest
Oscillatoria proteome has the longest protein with 3704 amino-acids: Ig-like domain-containing protein. That protein has several functions and one of them is antibacterial defense [5], which may be the exact reason for Oscillatoria to synthesize it. The shortest (28 amino-acids) is helix-turn-helix-domain-containing protein. This is a motif - short amino-acid sequence (talking about proteins) that hardly changes during evolution. Main function of that exact one is to bind the DNA and change the expression of genes. [6] Both proteins are located in the chromosome.
2.Genome
2.1 Start-codons
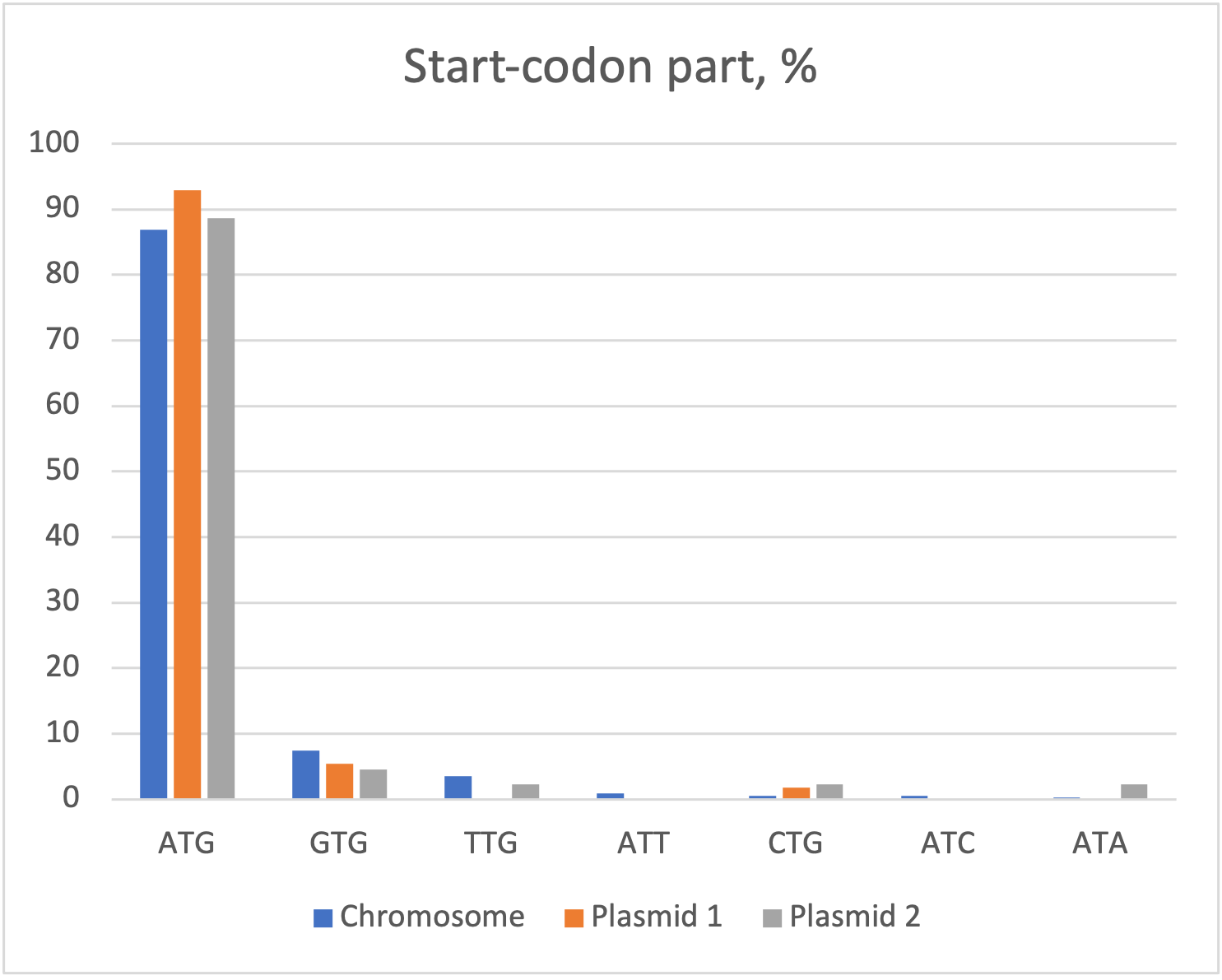
The most frequent one is ATG (86,96% of total). Then GTG (7,37%) and TTG with 3,52% (others part is under 1%).
2.2 GC-content
| GC-content,% | Chromosome | Plasmid 1 | Plasmid 2 |
|---|---|---|---|
| CDS | 48,23 | 47,73 | 49,74 |
| Between | 45,11 | 47,05 | 48,49 |
| All | 47,6 | 47,62 | 49,3 |
The given table (Table 2) shows the match between the data received from CDS and parts of sequence between them. So it is possible to predict the GC-content of CDS by their part between ORF that can also be justification of codon bias.[7]The difference in numbers in chromosome can be explained by the length of the sequence, which leads to a higher rate of uncertainties.
2.3 Stop-codons
| Stop-codon | Chromosome | Plasmid 1 | Plasmid 2 |
|---|---|---|---|
| TAA | 3298 | 30 | 26 |
| TAG | 1316 | 9 | 7 |
| TGA | 1137 | 17 | 11 |
As it can be seen from Table 3, TAA is the main stop-codon. According to the recent research [14], the main reason is reduction of translational readthrough (the phenomenon when stop-codon is perceived not as an end of translation, but as a sense-codon). According to the [8] TAA is mainly used in highly expressed proteins. In chromosome TAA stop-codon is mainly found in UMA2 family endonuclease, which is responsible for endonuclease activity [9], that makes internal breaks in nucleic acids [10]. Also in the proteins with tetratricopeptide repeat (TPR) motif, that are responsible for some vital processes in the cell [11]. And the last in top-3 is response regulator - signaling proteins, playing one of the leading roles in cell communication with the environment [12] In Plasmid 1 all the proteins with TAA stop-codon are in equal amounts. For example, there can also be found TPR proteins. Glycosyltransferases are involved in the synthesis of glycans - components of cell wall[13] Plasmid 2 has DNA/RNA non-specific endonuclease, site-specific integrase, involved into DNA binding, Rho termination factor N-terminal domain-containing protein, which function is to terminate the transcription [14]. So, I suggest that these proteins are highly expressed, according to the stop-codon they’re using and functions they have. This of course is not enough to call it a statement, so it’s just a suggestion.
Aknowledgments
I’d like to thank all the teachers of (bio)informatics who worked with us this term. It was really interesting, because that subject was the closest compared to others to the problems I hope I will encounter in the future. I appreciate Andrei Alekseevskiy, Ivan Rusinov, Arseni Zinkevich, Dmitry Penzar and Sergey Spirin.
References
[1] Wikipedia, Butylated hydroxytoluene
[2] Wikipedia, Oscillatoria
[3] Algae Base, Oscillatoria acuminata Gomont 1892
[4] Kerfeld C. A. et al (2012) Index of /genomes/all/GCF/000/317/105/GCF_000317105.1_ASM31710v1
[5]UniProt, Ig-like domain-containing protein - Homo sapiens (Human)
[6] Wikipdia, Helix-turn-helix
[7] Swaine L. Chen, William Lee, Alison K. Hottes, Lucy Shapiro, and Harley H. McAdams (2004) Codon usage between genomes is constrained by genome-wide mutational processes
[8] Gürkan Korkmaz, Mikael Holm, Tobias Wiens, Suparna Sanyal (2014) Comprehensive analysis of stop codon usage in bacteria and its correlation with release factor abundance
[9] UniProt, EWV80_17480 - Uma2 family endonuclease - Microcystis aeruginosa Ma_QC_B_20070730_S2
[10] Quick Go, Endonuclease activity
[11]Lukas Cerveny, Adela Straskova, Vera Dankova, Anetta Hartlova, Martina Ceckova, Frantisek Staud, and Jiri Stulike (2013) Tetratricopeptide Repeat Motifs in the World of Bacterial Pathogens: Role in Virulence Mechanisms
[12] Rong Gao, Timothy R. Mack, and Ann M. Stock (2007) Bacterial Response Regulators: Versatile Regulatory Strategies from Common Domains
[13] James M. Rini and Jeffrey D. Esko (2017) Essentials of Glycobiology [Internet]. 3rd edition. Chapter 6 «Glycosyltransferases and Glycan-Processing Enzymes»
[14]Alexander T Ho, Laurence D Hurst (2021) Variation in Release Factor Abundance Is Not Needed to Explain Trends in Bacterial Stop Codon Usage
[16] Wikipedia, Ribosomal RNA
[17] Text comparison
[18] Google Colaboratory