Homology and alignment
Description of carbohydrate binding domain
A carbohydrate-binding domain was selected for further work:
- AC: PF18448
- ID: CBM46
- Name: Carbohydrate binding domain
- Number of sequences in seed: 25
- Number of sequences in full: 243
- Number of architectures: 19 (the table from the task says only about 15)
Its function is to bind carbohydrates to facilitate their enzymatic hydrolysis (функция заключается в связывании углеводов, чтобы облегчить их ферментативный гидролиз). There are 19 (according to Pfam) architectures in this domain, the two largests have 169 and 52 proteins. There are two β-barrels in the 3D structure (was taken from Streptomyces hainanensis) predicted by AlphaFold.
Also there are 183 species with this domain, 183 of which are bacteria with 240 sequences and 2 is Fungi (Ascomycota and Basidiomycota) with 2 sequences. Interestingly enough that the findings are actually at opposite ends of the phylogenetic tree and can not be explained by the common origin of organisms. Horizontal gene transfer may have taken place but it is difficult to say for sure.
Multiple sequence alignment
Multiple sequence alignment (MSA) in fasta format sorted alphabetical can be found via link, sort according to tree is in the file.
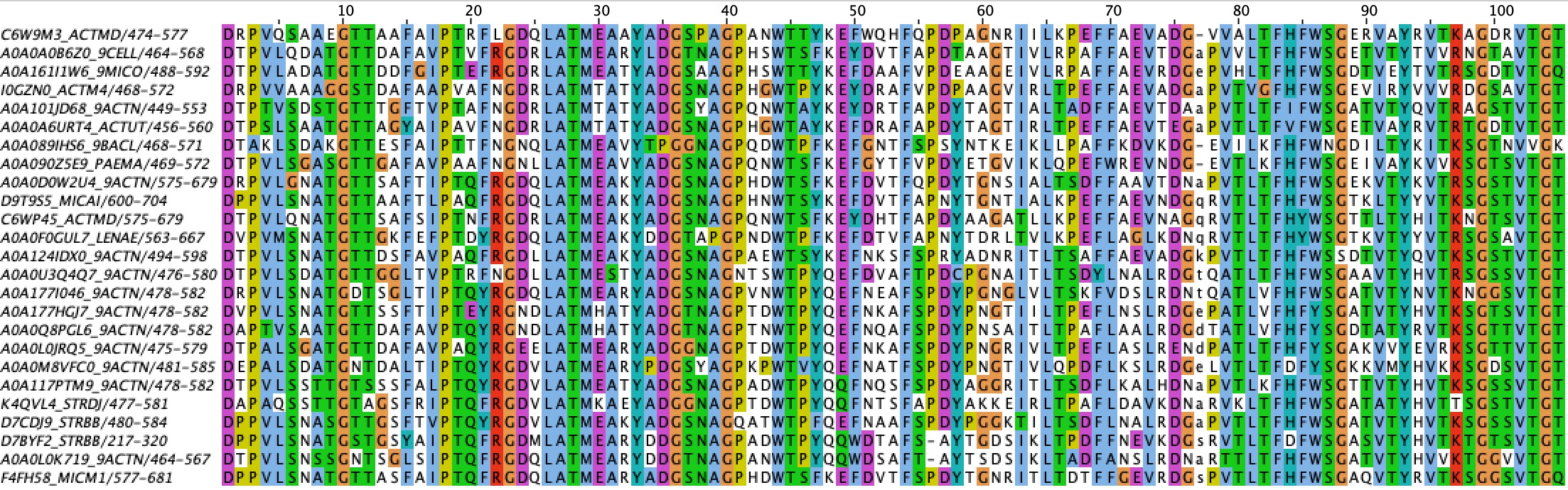
According to above identity threshold color scheme of MSA there are 20 columns with 100% identity (1, 10, 18, 23, 26-29, 33, 36, 40, 44-45, 54, 65, 83, 93, 99, 102, 104). The most conserved region is in columns 26-45 because identity is more than 90% in each column with the exception of column 32.
With above identity threshold color scheme of MSA with identity 0% there are the five least consreved regions: 13-14, 43-44, 52-53 and 59-63, their identity is less than 10% in each column. It does not shows the evolution and homology of sequences.
I think that 50-80 aa region is the most indicative in terms of homology because with a decrease in identity from 100% to 0% it has the biggest number of changes, which means it can well separate proteins by homology.