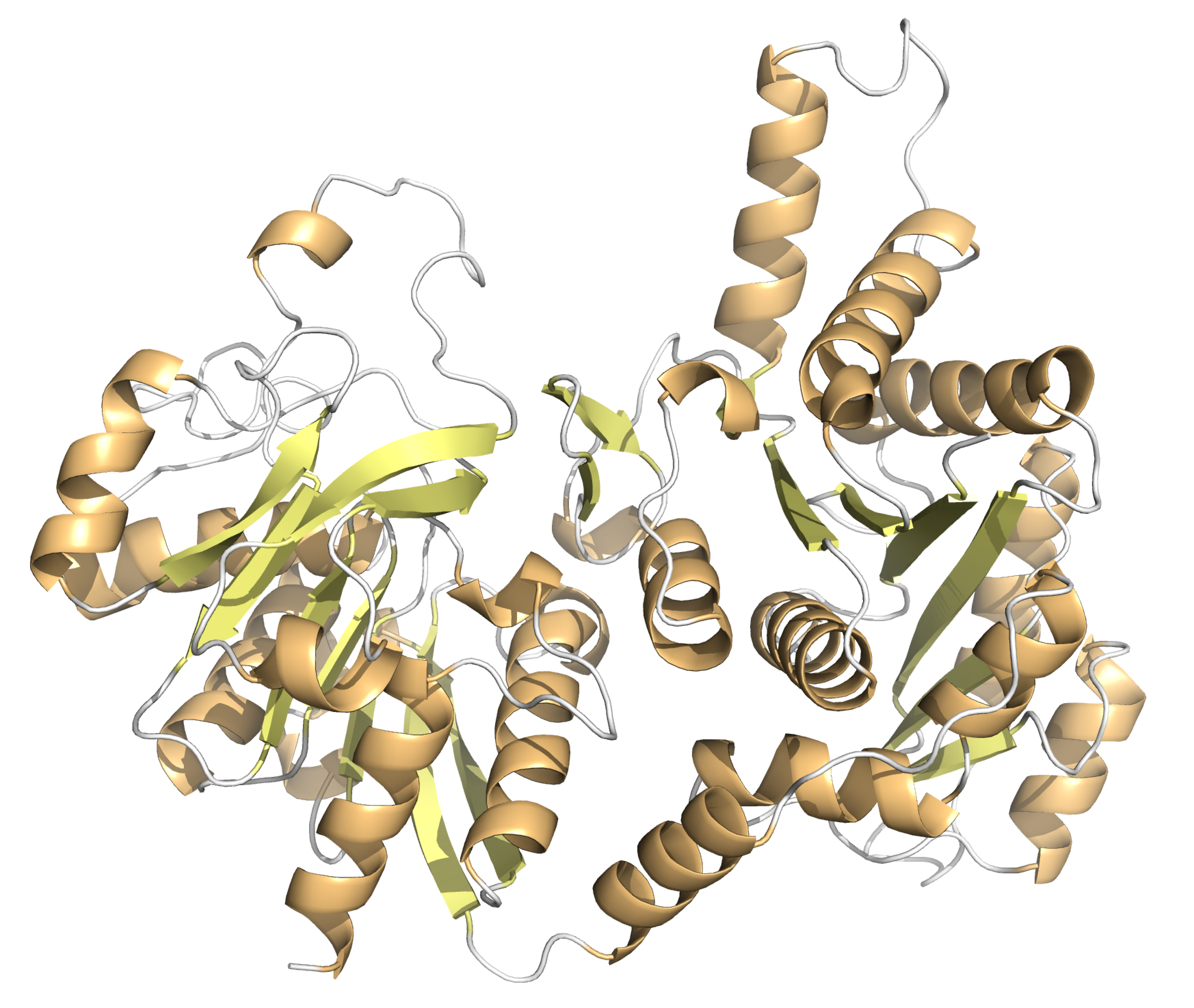
Description of the CTP synthase of archaea Thermococcus eurythermalis
Protein selection
I entered the name with (taxonomy_name:"Thermococcus eurythermalis") of my favorite archaea into the search in the UniRef database
and got a list of 2100 proteins.
I chose between well-annotated proteins: according to this parameter proteins with a rating of 4/5 are in the first place. There are only three proteins with this score: reverse gyrase, bifunctional NAD(P)H-hydrate repair enzyme and CTP synthase. So I deceided that the last one is the most interesting.
Protein information
The report file can be found at the link, also there is file with the information about protein clusters.
Based on description I understood that my favorite archaea uses CTP synthase (ЦТФ-синтаза, fig. 1), or cytidine 5'-triphosphate synthase for ATP-depended catalyzes of UTP amination (amine group from L-glutamine or nitrogenis introduced into UTP) forming CTP. So this protein regulates cellular levels of CTP through interactions with the four ribonucleotide triphosphates (ATP, UTP, CTP and GTP)
GTP is used for allosterically activation when the glutamine is the substrate of reaction (and in has no effect when ammonia is the substrate). So when there is a lot of GTP in the cell, the affinity of the enzyme to the substrate increases.
Search queries
Search: (ec:6.3.4.2), there are 56,557 results. CTP cynthase is found in most of the model organisms: yeast, bacteria
(including E. coli, Mycobacterium, photosynthetic Prochlorococcus etc.), Danio rerio, mice, cow and others including
humans (for them this protein is perfect annotated, 5/5).
I was wondering if it occurs in viruses, so I checked this fact with the help of a query (ec:6.3.4.2) AND (taxonomy_id:10239). So
thete areonly 11 proteins with annotation score 3/5 only for one viruse — Riboviria sp., the rest ten viruses have 2/5.
It is also interesting how many other proteins archaea have that use the CTF synthesized by my protein as a substrate. I used
(taxonomy_id:2157) AND (chebi:37563) ang got 12,227 results with lots of proteins like riboflavin kinase, CCA-adding enzyme,
uracil phosphoribosyltransferase and anothers (three mentioned are well-annotated, 5/5). I think it can be concluded that the substrate produced
by my protein is quite popular in archaea cells, which is to be expected. The frequency of using other substrates did not cause me any questions,
but to check how often exactly popular L-glutamine is used and got 47,055 results, for water (I think one of the most popular substrates) got
195,822 results.
But it's interesting to see how relevant this protein is to my archaea, so I used the query (taxonomy_id:1505907) AND
(keyword:"Pyrimidine metabolism") and got no results (the same for (taxonomy_id:1505907) AND
(keyword:"#Pyrimidine biosynthesis") from annotation of the protein). A further shortened query (taxonomy_id:1505907) AND
(keyword:"Pyrimidine") showed 9 finds. But the annotation score is no more than 3, often 2, so the usefulness of these proteins is
questionable.
Then I decided to change the database from UniProtKB to Proteomes and used the request (upid:UP000029980) AND (organism_id:1505907)
and got one proteome with full genome representation and 2,100 proteins (2,095 in chromosome and 5 in plasmid), most of which is single (99.1%),
next are missing with 0.9%.
References
CTP synthase page in UniProt (AC: A0A097QUU1_9EURY).