Results
1.Halapricum desulfuricans protein lengths histogram
Based on the protein data presented in CDS from the genome table of H.desulfuricans, the histogram of protein lengths was created. It reflects the features of length (number of amino acids in protein) distribution of proteins from H.desulfuricans proteome (Figure 1):
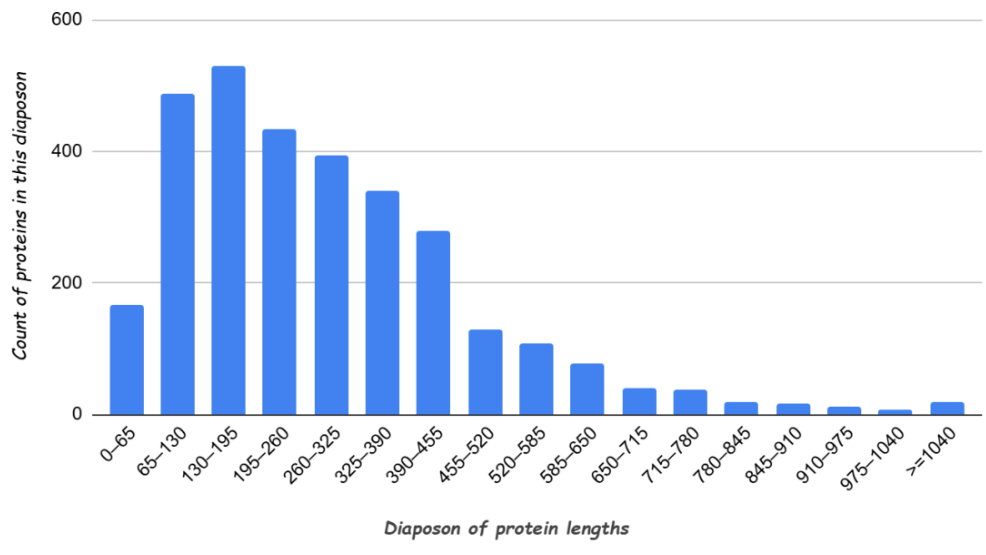
Figure 1. Histogram of H.desulfuricans protein lengths (minimal length – 24 aa, maximal length – 2384 aa).
Standard deviation of protein lengths is 608.937, coefficient of variation is 0.7015. As we can see, the spread of values of protein lengths is wide. Most proteins have length up to 455 amino acids. From the length of 455 amino acids number of proteins decreases. The largest number of proteins has length 130-195 amino acids.
The second place in number is occupied by proteins with lengths 65-130 amino acids, and the third place – by proteins with lengths 195-260 amino acids. Also, H.desulfuricans has a little number of proteins with length longer than 1040 amino acids, so I have decided to combine lengths from 1040 to 2384 in one range.
2.Halapricum desulfuricans intervals between CDS on + strand of chromosome lengths histogram
Based on the CDS data presented in the genomic features table of Halapricum desulfuricans, the histogram of interval between CDS on + strand of chromosome lengths was created. It shows the distribution of ranges of interval lengths between CDS on + strand of chromosome (Figure 2):
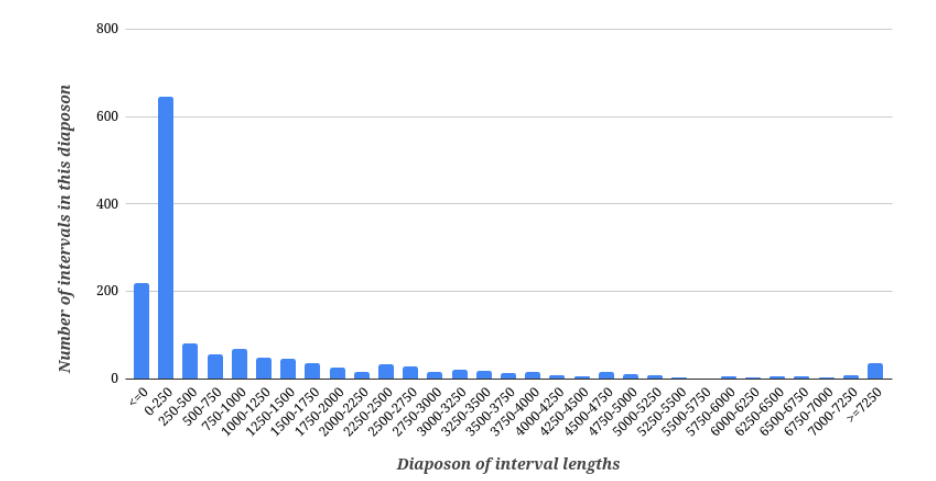
Figure 2. Histogram of H.desulfuricans interval between CDS on + strand of chromosome lengths (minimal length – -47 bp, maximal length – 12873 bp).
Standard deviation of interval lengths is 1920.157, coefficient of variation is 1.737. As we can see, the spread of values of interval lengths is wide, even wider than the protein lengths one. If the interval has length less than 0 base pairs, it means that the neighbouring CDS intersects. Most intervals have length up to 250 base pairs. The largest number of intervals has length 0-250 base pairs. The second place in number is occupied by intervals with lengths <=0 base pairs and the third place – by intervals with lengths 250-500 base pairs.
3.Halapricum desulfuricans intersections between CDS on + strand of chromosome lengths histogram
Based on the CDS data presented in the genomic features table of Halapricum desulfuricans, of intersection between CDS on + strand of chromosome lengths was created. It shows the distribution of ranges of intersection lengths between CDS on + strand of chromosome (Figure 3):
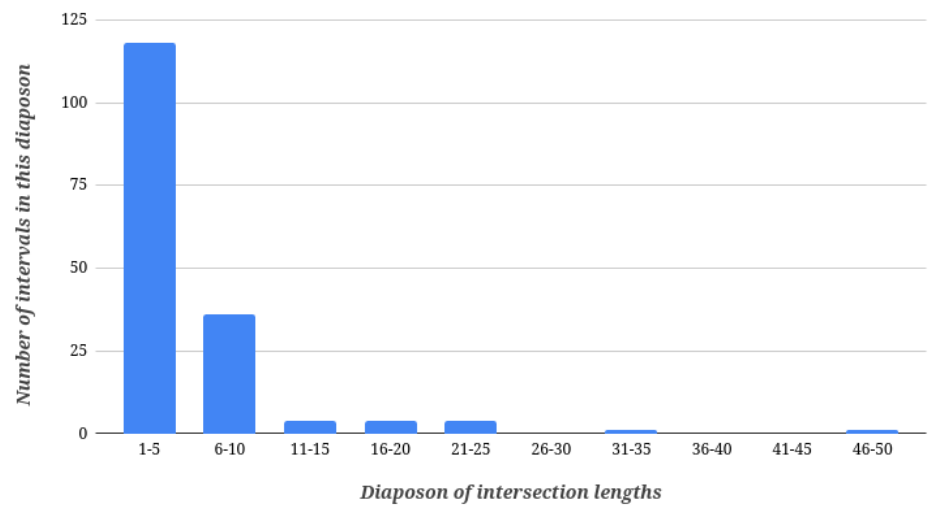
Figure 3. Histogram of H.desulfuricans intersection between CDS on + strand of chromosome lengths (minimal length – 1 bp, maximal length – 47 bp).
As we can see, the amount of intersections in Halapricum desulfuricans CDS is not huge. Also, lengths of intersections are not big: most intersections have length from 1 to 5 base pairs. Based on these data (paragraphs 2 and 3 from “Results”), we can conclude that for neighbouring CDS in Halapricum Desulfuricans genome, distances from 0 to 250 nucleotides are preferred and intersections are not preferred.
4. Per replicons of Halapricum desulfuricans genome count of protein coding genes and different types of RNA coding genes
Halapricum desulfuricans has two replicones: one chromosome and one plasmid. Chromosome includes most of bacteria’s genes (95,969% of Halapricum desulfuricans genome) and plasmid includes only 4,031% of Halapricum desulfuricans genome. Plasmid includes only protein coding genes and chromosome includes protein coding genes and different types of RNA coding genes. Most of the genome contains protein coding genes. There are 47 tRNA coding genes in the genome (Table 2). The number of different types of tRNA is less than the number of codons (61 without 3 stop codons) because of wobble base pairs – pairings between two nucleotides in RNA molecules that do not follow Watson-Crick base pair rules [3]. It expands the possible number of complementary interactions between tRNA and mRNA molecules [3]. There are 2 rRNA coding genes for each type of rRNA: one gene on each strand of chromosome.
Table 2. Per replicons of Halapricum desulfuricans genome count of protein coding genes and different types of RNA coding genes.
| Replicon | Count of proteins | Count of tRNA | Count of 23s rRNA | Count of 16s rRNA | Count of 5s rRNA | Percentage of the whole genome |
|---|---|---|---|---|---|---|
| Chromosome | 2899 | 47 | 2 | 2 | 2 | 95.969% |
| pHSR-Bgl01 | 124 | 0 | 0 | 0 | 0 | 4.031% |
5. Start codons in CDS count for pseudogenes and “normal” genes.
Table 3. Start codons in CDS of Halapricum desulfuricans count for pseudogenes and “normal” genes.
| Codons | All CDS | Pseudo CDS | Normal CDS |
|---|---|---|---|
| ATG | 2682 | 36 | 2646 |
| GTG | 321 | 7 | 314 |
| TTG | 41 | 0 | 41 |
| CTG | 10 | 0 | 10 |
| ATC | 9 | 1 | 8 |
| ATA | 4 | 1 | 3 |
| ATT | 2 | 1 | 1 |
| AAG | 1 | 1 | 0 |
| Others | 29 | 29 | 0 |
As we can see, Halapricum desulfuricans CDS has lots of start codon types (Table 3). Most of the start codons are ATG, GTG, TTG and CTG codons. In normal genes there are almost no other codons except for ATG, GTG, TTG and CTG, but in pseudogenes there is a wide variety of start codons. We can assume that one of the reasons for the transition of a "normal" gene into a pseudogene is the replacement of the start codon from NTG to some other. Also we can conclude that “normal” genes are much more common in the genome than pseudogenes.
6. Comparison of the GC-content of Halapricum desulfuricans genome and Escherichia coli genome
Table 4. Comparison of the GC-content of Halapricum desulfuricans genome and Escherichia coli genome.
| GC-percentage | Name |
|---|---|
| 63.85% | Halapricum desulfuricans |
| 56.00% | Escherichia coli |
As we can see, Halapricum desulfuricans genome contains 7,85% more GC (Table 4). Despite the fact that this archaea does not live in hot springs [2], it has a fairly high GC-content. It can be assumed that this feature was inherited from common ancestral Archaea which existed in conditions of high temperature.
7. Prediction of the quaternary structure of bacteriorhodopsin encoded in Halapricum desulfuricans genome.
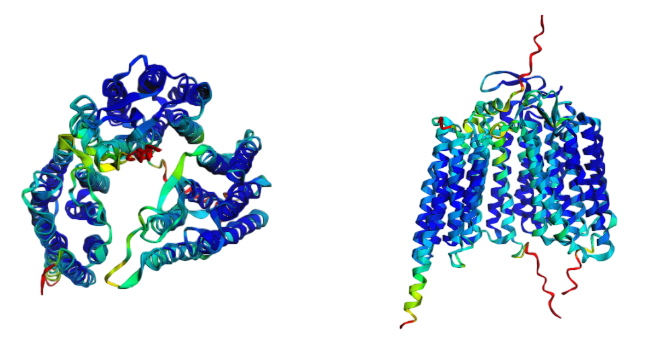
Figure 4. H.desulfuricans bacteriorhodopsin heterooligomer.
Since there is no structure of Halapricum desulfuricans bacteriorhodopsin in PDB, I have decided to predict its structure (Figure 4). Bacteriorhodopsin is a heteropolymer consisting of three polypeptide chains. It acts as a proton pump; that is, it captures light energy and uses it to move protons across the membrane out of the cell [4]. The proton-motive force generated by the protein is used by ATP synthase to generate ATP [4]. By expressing bacteriorhodopsin, the archaea cells are able to synthesise ATP in the absence of a carbon source.
Supplementary materials
Conclusion
In this work, Halapricum desulfuricans protein lengths histogram, Halapricum desulfuricans intervals between CDS on + strand of chromosome lengths histogram, Halapricum desulfuricans intersections between CDS on + strand of chromosome lengths histogram were created; per replicons of Halapricum desulfuricans genome count of protein coding genes and different types of RNA coding genes, start codons in CDS of Halapricum desulfuricans count for pseudogenes and “normal” genes were made; the GC-content of Halapricum desulfuricans genome and Escherichia coli genome were compared; the quaternary structure of bacteriorhodopsin encoded in Halapricum desulfuricans genome was predicted. I learned that archaeal genomes can contain significant diversity in start codons.
References
1. Parte, A.C., Sardà Carbasse, J., Meier-Kolthoff, J.P., Reimer, L.C. and
Göker, M. (2020). List of Prokaryotic names with Standing in
Nomenclature (LPSN) moves to the DSMZ. International Journal of
Systematic and Evolutionary Microbiology, 70, 5607-5612; DOI:
https://doi.org/10.1099/ijsem.0.004332
2. Dimitry Y. Sorokin, Michail M. Yakimov, Enzo Messina, Alexander Y. Merkel, Michel Koenen, Nicole J. Bale, Jaap S. Sinninghe Damsté. Halapricum desulfuricans sp. nov., carbohydrate-utilizing, sulfur-respiring haloarchaea from hypersaline lakes. Systematic and Applied Microbiology, Volume 44, Issue 6, November 2021. DOI: 10.1016/j.syapm.2021.126249
3. Cox, Michael M.; Nelson, David L. (2013). Lehninger Principles of Biochemistry (6th ed.). New York: W.H. Freeman. ISBN 978-0716771081.
4. Voet, Judith G.; Voet, Donald (2004). Biochemistry. New York: J. Wiley & Sons. ISBN 978-0-471-19350-0.