
According the task of pr 12 I used BLAST to search proteins which are potentionally homologous to my protein (0A0U3W9X5|A0A0U3W9X5_9BACI Fructose-1,6-bisphosphatase OS=Lentibacillus amyloliquefaciens). To search by similarity I chose database Swiss-Prot, algorothm protein-protein BLAST. When search settings were on default (E-value = 10, WordSize = 6, Max target sequences = 100) 36 proteins were found and only one with large e-value, so then I set E-value = 20 to find two proteins with large e-value (37 proteins were found).
Table 1. Characteristics 9 selected findings of the BLAST.
*Line with "my protein" is in bold.
| # | ID/AC | Name of protein | Coverage | Identity % | E-value | Homologous |
| 1 | A0A0U3W9X5_9BACI/A0A0U3W9X5 | Fructose-1,6-bisphosphatase | ||||
| 2 | GLPX_BACSU/Q03224.1 | Fructose-1,6-bisphosphatase class 2 | 100% | 74% | 3e-174 | yes |
| 3 | GLPX_CORGL/Q6M6E7.1 | Fructose-1,6-bisphosphatase class 2 | 97% | 55% | 1e-115 | yes |
| 4 | GLPX_MYCBO/Q7U0N5.2 | Fructose-1,6-bisphosphatase class 2 | 97% | 51% | 2e-111 | yes |
| 5 | FBSB_CYAA5/B1WQ07.1 | D-fructose 1,6-bisphosphatase class 2 | 99% | 48% | 2e-109 | yes |
| 6 | FBSB_MICAN/B0JKN5.1 | D-fructose 1,6-bisphosphatase class 2 | 99% | 47% | 3e-109 | yes |
| 7 | GLPX_MYCLB/B8ZSG6.2 | Fructose-1,6-bisphosphatase class 2 | 97% | 50% | 1e-107 | yes |
| 8 | FBSB2_ACAM1/B0BZF8.1 | D-fructose 1,6-bisphosphatase class 2 | 98% | 48% | 1e-106 | yes |
| 9 | RDRP_PVMR/P95469.1 | RNA replication polyprotein | 15% | 39% | 2.8 | not |
| 10 | RECA_PARDE/P95469.1 | Protein RecA | 25% | 30% | 19 | not |
Picture 1. The representation of alignment. (You can also see it here)

Analysis of the homology of finding with small e-value
Analysis of the homology of finding with large e-value.
For this task I used Pfam where I searched proteins which are included a family ZZ. I chose two proteins with the following architectures whose differences seemed interesting but not very drastically to me. These protein's ID you can see in a table 2.
Table 2. Proteins having homologous sections (domains).
| Architecture | ID/AC | Name of the protein | Organism |
| ZZ, N_BRCA1_IG | A0A0D2CWB4_9EURO/A0A0D2CWB4 | Uncharacterized protein | Exophiala xenobiotica |
| PB1, ZZ, N_BRCA1_IG | F4PSP5_DICFS/F4PSP5 | ZZ-type zinc finger-containing protein | Dictyostelium fasciculatum (strain SH3) |
For searching I set E-value = 1.0E-5, WordSize = 2 and I didn't change other settings so they were on default.
Picture 2. Map of the local similarity.
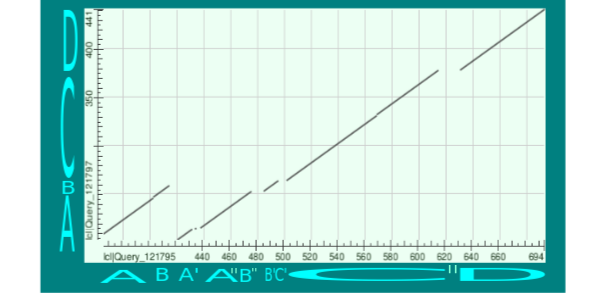
Analysis of the map of the local similarity.
Let's look at vertical axis which shows the query sequence (A0A0D2CWB4). We can see 4 parts of the homologous units: A, B, C, D. Then look at horizontal axis which shows the database sequence (F4PSP5). It compared of the same parts with with some modifications.
There is duplication of parts A and B in the database sequence. And there are some deletions (or nonhomologous inserts - ?) in the second duplicated unit. The deletions cut the second unit A to A' and A'', the second unit B to B' and B'' and the unit C to C' and C''. Units B'' and C' aren't divided, but I called them in different ways because they are visual divided if you look at query sequence.