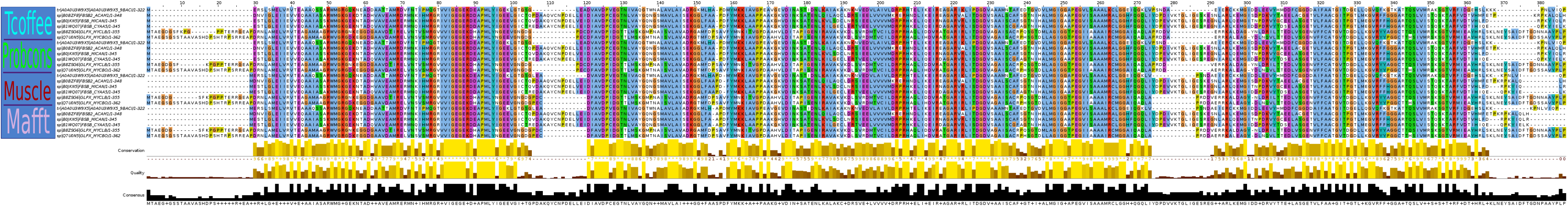
According the task I used 5 homologous proteins which I found in pr12. You can see them characteristics in the table 1. Then I aligned them by programs Tcoffee, Probcons, Muscle and Mafft. Concatenating the result of the alignmenst you can see on picture 1.
Table 1. Characteristics of homologous proteins which were used.
*Line with "my protein" is in bold.
| # | ID/AC | Name of protein | Coverage | Identity % | E-value |
| 1 | A0A0U3W9X5_9BACI/A0A0U3W9X5 | Fructose-1,6-bisphosphatase | |||
| 4 | GLPX_MYCBO/Q7U0N5.2 | Fructose-1,6-bisphosphatase class 2 | 97% | 51% | 2e-111 |
| 5 | FBSB_CYAA5/B1WQ07.1 | D-fructose 1,6-bisphosphatase class 2 | 99% | 48% | 2e-109 |
| 6 | FBSB_MICAN/B0JKN5.1 | D-fructose 1,6-bisphosphatase class 2 | 99% | 47% | 3e-109 |
| 7 | GLPX_MYCLB/B8ZSG6.2 | Fructose-1,6-bisphosphatase class 2 | 97% | 50% | 1e-107 |
| 8 | FBSB2_ACAM1/B0BZF8.1 | D-fructose 1,6-bisphosphatase class 2 | 98% | 48% | 1e-106 |
Picture 1. The representation of multiple alignment. (You can also see it here)
*You can see list of programs which were used for alignments in the left column.

Differences between programs Tcoffee and Probcons:
Sum up, there are minor differences between the alignments. Overall they are very similar. I think Probcons' alignments might carry more weight than Tcoffee's alignment.
I used Pfam to search for my protein's architecture. Then I found other proteins including the same domain but which have some differences in architecture.
Description of the architecture of my protein:
There is only one domain in my protein: FBPase_glpX (See picture 2, table 2). It includes the part of sequence from the third residue to the 310th residue (the lenght of the whole sequence is 322 residues).
Picture 2. Domain organisation of the Fructose-1,6-bisphosphatase.

Table 2. Some data about FBPase_glpX.
| Source | Domain | Start | End |
| Pfam | FBPase_glpX | 3 | 310 |
| disorder | n/a | 20 | 32 |
| disorder | n/a | 34 | 41 |
| disorder | n/a | 79 | 80 |
| low_complexity | n/a | 141 | 152 |
Some other architectures including FBPase_glpX domain (see pictures 3, 4, 5, 6):
Picture 3.

Picture 4.

Picture 5.

Picture 6.
