| FBB MSU site | Main page | About me | Terms |
Alignment of the genomes
Task 1 and Task 3 / Part 1
I chose three genomes of bacteria belonging to the same species (Acinetobacter baumannii) which have one chromosome.
- Acinetobacter baumannii strain SAA14 (CP020579.1), Complete Genome
- Acinetobacter baumannii strain AB042 (CP019034.1), Complete Genome
- Acinetobacter baumannii strain AB031 (CP009256.1), Complete Genome
As you can see, these sequences have large sections of homologous and contain large genomic rearrangements. Here are dot matrix view of pairvise alignment made by blast 2 sequences* (BLASTN 2.8.0) and them analysis - see fig. 1-3.
*I used Word size = 256.
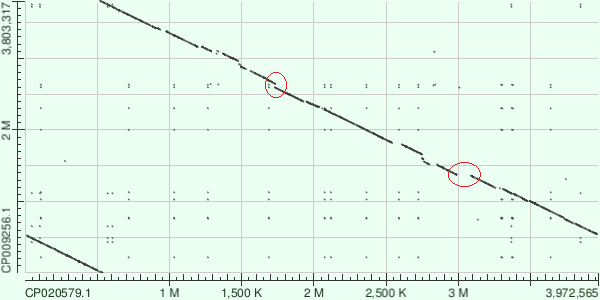
| Figure 1. SAA14 vs AB031. Alignment. |  |
| Sequences start from different places. Deletions or inserts are marked in red. | |
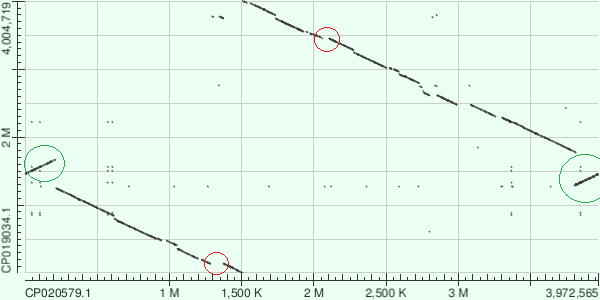
| Figure 2. SAA14 vs AB042. Alignment. |  |
| Sequences start from different places. Deletions or inserts are marked in red and inversionsions are marked in green. | |
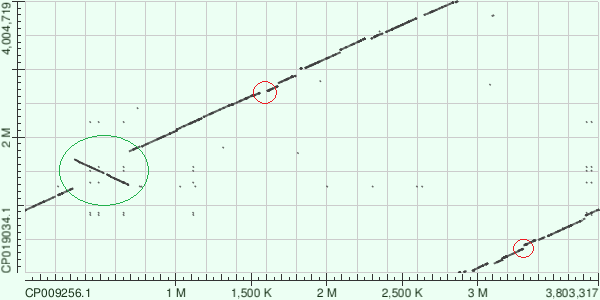
| Figure 3. AB031 vs AB042. Alignment. | 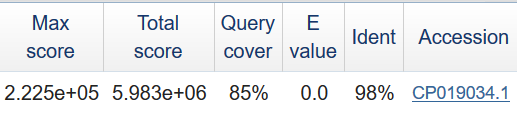 |
| equences start from different places. Deletions or inserts are marked in red and inversionsions are marked in green. | |
Task 2
According the instruction I created file genomes.tsv for NPG-explorer with references to FASTA files contains sequenses of genomes of chosen bacterias. Then I run npge MakePangenome and PostProcessing (protocol of it's work) and visualised the result by qnpge.
The result of searching you can see in table 1. To complete this table I used data from pangenome.info. The summary length of blocks is 37056887, the summary cover of blocks is 74.98%.
Table 1.
| All non-minor blocks of at least 2 fragments | S-blocks | H-blocks | R-blocks | M-blocks | |
| Number | 941 | 338 | 305 | 298 | 434 |
| Block identity | min=0.7692 median=0.952 avg=0.9374 max=1 | min=0.812 median=0.9678 avg=0.9515 max=1 | min=0.8017 median=0.9633 avg=0.948 max=1 | min=0.7692 median=0.8951 avg=0.9107 max=1 | min=0 median=0.8492 avg=0.7162 max=1 |
| Identity of joined blocks | 0.971479 | 0.973221 | 0.965706 | 0.94722 | 0.877436 |
| Length and cover of fragments | 10557660 (89.61%) | 9242570 (78.45%) | 726889 (6.17%) | 588201 (4.99%) | 24492 (0.2%) |
| Total length and cover of blocks | 3581022 (74.73%) | 3082501 (64.32%) | 363773 (7.59%) | 134748 (2.81%) | 12465 (0.26%) |
Task 3 / Part 2
To find the large genomic rearrangements I used qnpge-visualisation, blocks.gbi, blocks.blocks. I found out that there are 33 of g-blocks and 64 of i-blocks. Below you can see some examples of rearrangements.
Figure 4. Inversion of the fragment g3x157172-i2x15531-g3x200780 in AB042 (see positions 10-12).

Figure 5. Translocation of g3x102 in SAA14 (see position 54 and 58).

Figure 6. Conserved blocks in positions 40 and 44. Insert of a fragment between them in AB042.

Phylogeny
The phylogenetic tree made by Blast Tree View (see fig. 7).
Figure 7.

| Term 3 |
| ← Pr 9 | Block 3 → |
© Darya Potanina, 2017