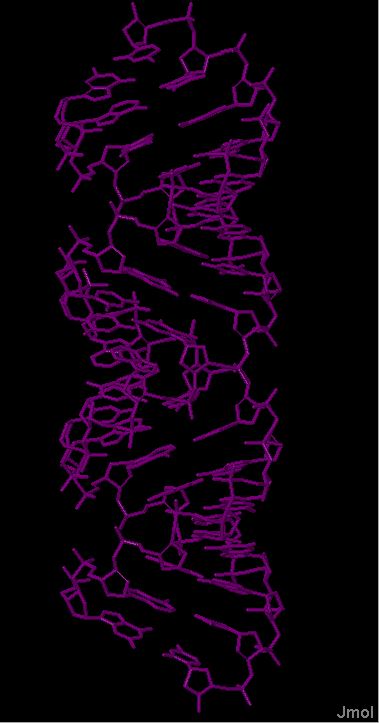
Построение моделей структур А-, В- и Z-формы ДНК с помощью инструментов пакета 3DNA.
Работа в JMol со структурами нуклеиновых кислот.

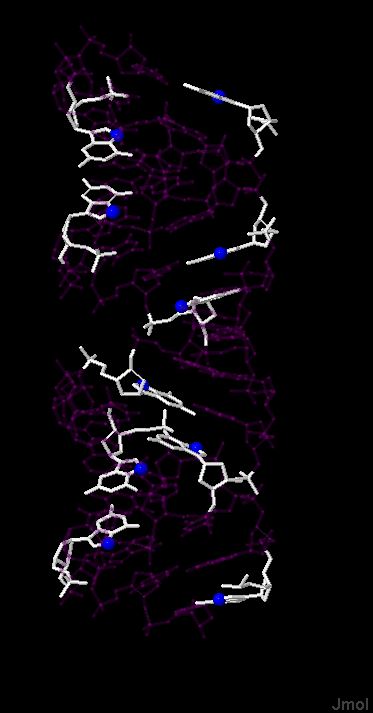
Проверка структур ДНК и РНК на наличие разрывов.
Сравнение скруктур трех форм ДНК с помощью средств JMol.
| А-форма | В-форма | Z-форма | |
| Тип спирали (правая или левая) | правая | правая | левая |
| Шаг спирали | 28,03 А | 33,75 А | 43,5 А |
| Число оснований на виток | 11 | 10 | 12 |
| Ширина большой борозды | 16,81 A (G33:B.P-A10:A.P) | 17,21 A (C4:A.P-A34:B.P) | 18,3 A (C56:B.P-C22:A.P) |
| Ширина малой борозды | 7,98 A (A10:A.P-G25:B.P) | 11,69 A (G33:B.P-C12:A.P) | 7,2 A (G17:A.P-G67:B.P) |
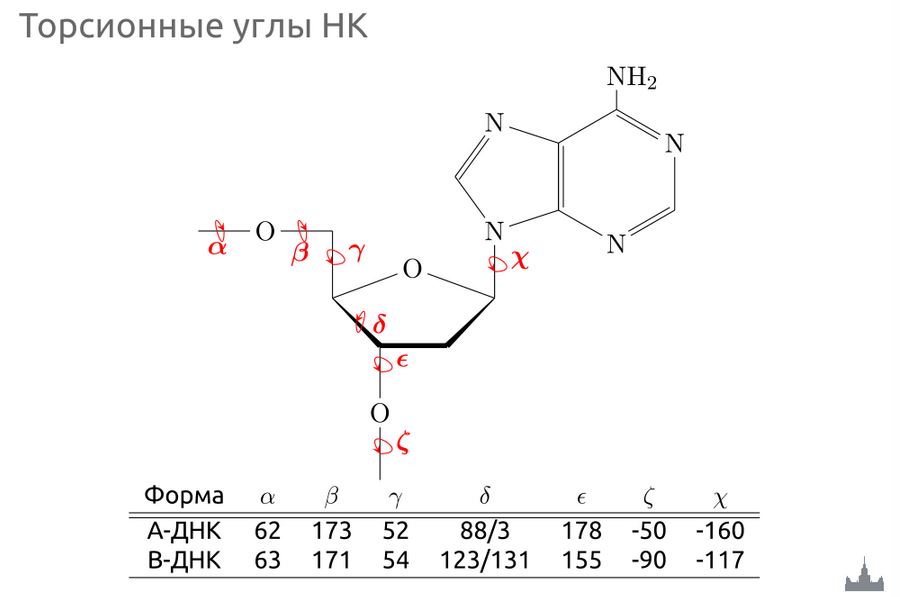
Сравнение торсионных углов в А- и В-формах белка
| Форма | ? | ? | ? | ? | ? | ? | ? |
| А-ДНК | 64.1 | 174.8 | 41.7 | 79.1 | -147.8 | -75.1 | -156.5 |
| В-ДНК | 85.9 | 136.3 | 31.1 | 143.3 | -140.8 | -160.5 | -98.5 |
Определение параметров структур нуклеиновых кислот с помощбю программ пакета 3DNA
Определим сначала торсионные углы для А-, В- и Z-форм ДНК.
A-форма Strand I base alpha beta gamma delta epsilon zeta chi 1 G --- 174.8 41.7 79.0 -147.8 -75.1 -157.2 2 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 3 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 4 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 5 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 6 A -51.7 174.8 41.7 79.0 -147.8 -75.1 -157.2 7 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 8 C -51.7 174.8 41.7 79.0 -147.8 -75.0 -157.2 9 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 10 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 11 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 12 C -51.7 174.8 41.7 79.1 -147.7 -75.1 -157.2 13 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 14 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 15 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 16 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 17 G -51.7 174.8 41.7 79.0 -147.8 -75.1 -157.2 18 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 19 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 20 C -51.7 174.8 41.7 79.1 --- --- -157.2 Strand II base alpha beta gamma delta epsilon zeta chi 1 C -51.7 174.8 41.7 79.0 --- --- -157.2 2 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 3 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 4 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 5 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 6 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 7 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 8 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 9 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 10 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 11 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 12 G -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 13 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 14 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 15 A -51.7 174.8 41.7 79.0 -147.8 -75.1 -157.2 16 G -51.7 174.8 41.7 79.1 -147.7 -75.1 -157.2 17 C -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 18 T -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 19 A -51.7 174.8 41.7 79.1 -147.8 -75.1 -157.2 20 G --- 174.8 41.7 79.1 -147.8 -75.1 -157.2 B-форма Strand I base alpha beta gamma delta epsilon zeta chi 1 G --- 136.4 31.1 143.4 -140.8 -160.5 -98.0 2 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 3 T -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9 4 C -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 5 G -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 6 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 7 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 8 C -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9 9 G -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 10 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 11 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 12 C -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 13 G -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0 14 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 15 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 16 C -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 17 G -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 18 A -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0 19 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 20 C -29.9 136.3 31.2 143.3 --- --- -98.0 Strand II base alpha beta gamma delta epsilon zeta chi 1 C -29.9 136.4 31.1 143.4 --- --- -98.0 2 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 3 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 4 G -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0 5 C -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 6 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 7 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 8 G -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 9 C -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9 10 T -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 11 A -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 12 G -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 13 C -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 14 T -29.9 136.3 31.1 143.3 -140.8 -160.5 -97.9 15 A -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 16 G -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 17 C -29.9 136.3 31.2 143.3 -140.8 -160.5 -98.0 18 T -29.9 136.4 31.1 143.4 -140.8 -160.5 -98.0 19 A -29.9 136.3 31.1 143.3 -140.8 -160.5 -98.0 20 G --- 136.3 31.2 143.3 -140.8 -160.5 -98.0 Z-форма Strand I base alpha beta gamma delta epsilon zeta chi 1 G --- 179.0 -173.8 94.9 -103.6 -64.8 58.7 2 C -139.5 -136.7 50.9 137.6 -96.5 81.9 -154.3 3 G 52.0 179.0 -173.8 94.9 -103.6 -64.8 58.7 4 C -139.5 -136.8 50.8 137.6 -96.5 82.0 -154.3 5 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7 6 C -139.5 -136.8 50.9 137.6 -96.5 82.0 -154.3 7 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7 8 C -139.5 -136.7 50.9 137.6 -96.5 81.9 -154.3 9 G 52.0 179.0 -173.8 94.9 -103.6 -64.8 58.7 10 C -139.5 -136.8 50.8 137.6 --- --- -154.3 Strand II base alpha beta gamma delta epsilon zeta chi 1 C -139.5 -136.7 50.9 137.6 --- --- -154.3 2 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7 3 C -139.5 -136.8 50.9 137.6 -96.5 82.0 -154.3 4 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7 5 C -139.5 -136.8 50.8 137.6 -96.5 82.0 -154.3 6 G 52.0 179.0 -173.8 94.9 -103.6 -64.8 58.7 7 C -139.5 -136.7 50.9 137.6 -96.5 81.9 -154.3 8 G 51.9 179.0 -173.8 94.9 -103.6 -64.8 58.7 9 C -139.5 -136.8 50.9 137.6 -96.5 82.0 -154.3 10 G --- 179.0 -173.8 94.9 -103.6 -64.8 58.7
Можно заметить, что при различных основаниях во всех структурах углы практически совпадают. Поэтому можно составить более компактную таблицу.
| Форма | ? | ? | ? | ? | ? | ? | ? |
| А-ДНК | -51.7 | 174.8 | 41.7 | 79 | -147.8 | -75.1 | -157.2 |
| В-ДНК | -29.9 | 136.35 | 31.15 | 143.3 | -140.8 | -160.5 | -98 |
| Z-ДНК (для цитозинов) | -139.5 | -136.75 | 50.85 | 137.6 | -96.5 | 82 | -154.3 |
| Z-ДНК (для гуанинов) | 51.9 | 179 | -173.8 | 94.9 | -103.6 | -64.8 | 58.7 |
Посмотрев на таблицу, можно сделать вывод:
Торсионные углы для тРНК 1FMT.
Strand I base alpha beta gamma delta epsilon zeta chi 1 G --- 134.7 75.1 79.1 -135.0 -86.2 177.3 2 C -49.2 173.5 45.7 81.5 -136.8 -68.5 -151.1 3 G -57.5 169.1 58.9 78.3 -165.9 -82.5 -171.4 4 G -70.1 179.0 59.4 86.2 -150.9 -64.8 -173.7 5 G -69.5 177.8 64.5 81.9 -167.1 -76.1 -176.9 6 G -77.3 -172.7 61.3 92.9 --- --- -161.6 7 G --- -125.1 76.7 89.5 -160.7 -63.6 179.8 8 U -74.6 -165.7 54.3 87.8 -159.7 -67.1 -146.4 9 C -71.3 -175.4 45.3 80.4 -150.7 -73.7 -152.3 10 G -69.6 157.9 66.5 79.4 -158.8 -69.8 -170.3 11 G -69.0 -170.5 53.3 80.3 -163.5 -66.2 -157.7 12 u -65.6 -167.7 48.4 82.7 -145.5 -48.8 -153.9 13 P -71.5 -175.5 52.4 88.0 --- --- -124.7 14 A --- 167.9 55.4 78.2 --- --- -167.9 15 A --- -116.9 58.1 87.4 -167.8 -65.4 178.1 16 C -68.9 -170.2 52.5 87.3 -156.8 -71.9 -157.4 17 C -53.8 174.4 40.4 79.9 -157.3 -72.4 -167.5 18 C -66.7 176.8 51.0 80.9 -150.6 -72.8 -170.8 19 G -58.0 174.3 53.3 80.7 -156.7 -70.9 -165.3 20 A -61.2 174.9 55.8 82.0 -151.3 -91.2 -162.6 21 A -39.6 157.2 45.9 84.4 --- --- -148.6 22 G --- -168.5 54.5 83.6 -156.3 -65.6 178.0 23 A -71.6 -162.1 46.9 83.3 -159.7 -70.5 -167.5 24 G -72.5 -172.7 51.8 83.7 -166.0 -53.8 -159.1 25 C 167.7 -179.9 171.8 86.3 -151.0 -64.4 -165.7 26 A -72.0 -175.5 56.8 77.8 -157.3 -54.6 -170.9 27 G -71.4 176.6 63.5 89.3 --- --- -157.6 28 G --- -172.8 157.3 148.1 --- --- -105.6 29 G --- -163.7 61.3 81.8 -147.6 -77.2 -171.4 30 C -57.8 -175.0 42.9 79.9 -139.1 -68.7 -150.9 31 G -58.8 168.3 63.4 78.1 -159.1 -86.6 -171.1 32 G -59.8 170.5 58.0 86.8 -150.2 -63.8 -171.0 33 G -74.7 178.2 62.5 83.8 -173.9 -71.8 -173.8 34 G -85.5 -162.3 64.8 98.5 --- --- -158.7 35 G --- -120.4 73.4 89.9 -164.6 -60.3 -179.9 36 U -75.5 -163.6 53.8 86.7 -152.2 -73.6 -151.7 37 C -65.9 177.5 44.7 81.2 -153.1 -75.6 -148.9 38 G -63.6 155.2 61.2 77.9 -157.5 -70.0 -169.1 39 G -66.2 -173.2 52.4 81.1 -160.7 -67.8 -161.2 40 u -69.6 -167.0 48.7 81.8 -149.6 -47.3 -151.6 41 P -82.4 -166.9 55.2 91.0 --- --- -134.4 42 A --- 179.6 49.7 86.1 --- --- -156.2 43 A --- -155.6 51.0 81.5 -144.5 -70.5 -168.6 44 C -72.5 -172.2 49.1 86.0 -151.9 -70.6 -157.7 45 C -61.4 175.2 51.1 83.1 -163.6 -72.8 -166.2 46 C -73.8 -177.2 52.1 82.6 -156.3 -73.9 -168.1 47 G -58.5 178.0 52.0 84.6 -157.0 -71.9 -165.9 48 A -56.7 176.7 51.0 84.2 -152.5 -88.1 -164.5 49 A -46.1 159.3 50.8 85.2 --- --- -145.5 50 G --- -163.7 52.4 85.3 -151.2 -68.8 177.7 51 A -68.5 -167.3 51.2 84.2 -158.6 -69.1 -168.4 52 G -79.2 -169.4 52.8 81.3 -165.2 -36.3 -154.9 53 C -168.4 156.6 162.8 85.6 -150.9 -64.0 -164.6 54 A -70.4 -174.4 52.4 78.4 -155.5 -57.7 -171.6 55 G -67.5 174.5 62.7 88.7 -126.4 -56.5 -157.1 56 C -75.9 -157.0 54.1 149.4 --- --- -81.9 57 G --- -115.2 -171.5 146.8 --- --- -141.2 Strand II base alpha beta gamma delta epsilon zeta chi 1 C -48.4 167.0 56.7 81.3 --- --- 178.5 2 G -71.8 -165.7 51.2 82.8 -145.2 -77.7 -178.5 3 C -64.6 176.5 55.0 82.4 -175.9 -71.0 -165.8 4 C -68.6 -172.0 45.2 80.9 -159.8 -73.0 -175.1 5 C -58.6 179.6 51.9 83.9 -159.5 -69.4 -157.8 6 C -68.0 177.6 53.6 82.7 -153.8 -65.8 -158.3 7 C -54.1 170.8 55.9 82.0 -149.1 -80.3 -165.5 8 G -76.3 -162.9 48.8 81.8 -158.1 -67.4 -162.9 9 G -68.2 -179.7 51.9 81.8 -168.1 -66.9 -160.2 10 C -73.1 -164.6 45.4 82.8 -157.5 -74.1 -160.2 11 C --- -178.2 40.1 82.2 -151.9 -70.5 177.0 12 A --- -153.9 53.9 147.4 --- --- -88.5 13 G --- -64.2 -155.3 147.8 --- --- 175.6 14 U 177.4 148.4 143.6 87.8 --- --- 177.5 15 c -45.4 169.0 46.4 79.9 172.8 -22.3 -157.4 16 G -56.5 -172.2 43.8 78.7 -145.5 -67.2 -162.7 17 G -51.8 164.2 55.1 77.2 -155.8 -63.9 -170.5 18 G -54.0 175.9 45.3 79.8 -146.8 -74.1 -164.6 19 C -62.7 165.8 46.8 79.5 -151.3 -77.9 -147.6 20 U -59.4 171.2 48.5 82.3 -136.3 -83.1 -168.9 21 G -64.6 178.7 47.8 83.3 -148.8 -66.3 -173.2 22 C 148.0 -171.7 -175.4 85.1 -147.0 -67.2 -178.6 23 U -69.4 179.3 51.1 83.8 -167.2 -60.1 -165.5 24 C -56.8 -177.5 47.1 83.9 -148.6 -70.5 -159.2 25 G --- 135.1 36.9 78.9 -155.3 -70.0 -177.2 26 u --- 158.6 54.7 84.5 --- --- -173.8 27 C --- -101.4 167.0 142.1 --- --- -164.6 28 C --- 166.7 47.5 85.6 --- --- -153.3 29 C -51.4 173.3 58.7 83.5 --- --- 179.4 30 G -85.8 -154.9 54.3 82.5 -150.3 -74.6 -179.5 31 C -61.8 169.0 55.9 83.4 174.9 -65.6 -167.0 32 C -62.4 -174.8 43.0 82.1 -154.8 -77.4 -173.4 33 C -60.4 -178.4 52.9 85.1 -159.2 -72.3 -161.4 34 C -65.1 179.0 51.1 81.4 -149.5 -67.0 -157.2 35 C -53.3 173.0 55.4 80.0 -152.9 -74.0 -159.2 36 G -71.7 -168.3 45.1 79.9 -159.0 -66.7 -162.9 37 G -67.7 176.6 51.3 83.6 -162.8 -69.7 -164.1 38 C -75.3 -160.8 43.3 81.8 -155.4 -76.9 -159.8 39 C --- 177.0 39.8 82.4 -157.6 -63.0 -178.5 40 A --- -156.5 55.9 145.4 --- --- -95.4 41 G --- 138.6 145.2 148.9 --- --- -160.4 42 U -92.0 -176.5 64.5 80.6 --- --- -163.1 43 c -38.7 159.6 40.7 79.4 -159.9 -54.2 -161.1 44 G -50.2 -179.6 43.5 78.5 -143.0 -72.6 -161.4 45 G -59.9 164.3 61.5 80.2 -160.4 -68.9 -171.4 46 G -52.6 177.8 44.3 81.6 -145.4 -73.1 -163.1 47 C -66.4 169.6 47.7 81.6 -154.5 -76.1 -150.2 48 U -68.2 172.2 59.1 80.5 -134.7 -80.8 -172.7 49 G -66.6 177.6 50.8 83.1 -149.8 -66.4 -167.6 50 C 152.2 -171.2 -176.5 83.4 -141.7 -66.8 176.3 51 U -74.4 -175.8 52.4 85.4 -167.0 -62.3 -158.8 52 C -51.0 176.6 47.5 82.1 -147.4 -71.4 -169.7 53 G --- 142.7 33.3 80.0 -154.9 -74.0 -178.8 54 u --- 160.5 53.3 83.2 --- --- -176.9 55 C --- 99.9 71.2 135.2 --- --- -155.5 56 A --- 167.9 59.7 87.7 --- --- 178.8 57 C --- 153.0 38.6 82.7 --- --- -155.5
Трудно определить на какую форму ДНК похожа это молекула тРНК. Вероятно, больше всего похожа на Z-форму.
Торсионные углы DNA-protein complex 1DC1
Strand I base alpha beta gamma delta epsilon zeta chi 1 T --- 148.3 84.9 87.9 172.7 -71.2 -128.3 2 A -59.2 176.1 48.2 139.5 -64.7 146.8 -106.8 3 C 57.2 -143.1 -171.1 82.6 -155.1 -68.4 -163.7 4 T -77.0 -174.1 54.2 84.9 -145.8 -62.8 -154.5 5 C -64.0 -179.9 60.1 139.5 -147.4 -160.3 -115.7 6 G -49.4 145.1 53.9 151.5 -89.4 -172.9 -107.3 7 A -100.9 139.9 39.1 73.1 -168.5 -76.1 -147.3 8 G 158.4 -151.7 170.2 98.4 -153.6 -66.4 -171.2 9 T -61.6 176.1 54.5 95.4 -173.5 -74.2 -155.1 10 A -57.7 -177.7 56.3 138.6 -151.0 -115.6 -116.6 11 T -59.0 155.8 50.9 131.3 --- --- -100.1 Strand II base alpha beta gamma delta epsilon zeta chi 1 A -44.4 -179.8 44.4 139.2 --- --- -114.0 2 T -66.6 178.4 53.2 90.7 -171.4 -81.1 -151.2 3 G 153.2 -149.9 173.2 97.4 -152.6 -66.5 -175.9 4 A -100.9 142.4 37.6 76.3 -173.2 -77.5 -145.9 5 G -49.8 147.3 53.2 149.5 -87.7 -174.7 -108.3 6 C -59.5 179.1 58.1 140.2 -149.4 -156.6 -116.7 7 T -71.4 -172.6 50.3 84.7 -145.8 -65.4 -150.5 8 C 60.4 -150.8 -173.9 86.6 -155.0 -68.3 -161.9 9 A -41.7 163.3 39.0 134.1 -60.5 142.2 -105.6 10 T -43.8 167.8 43.0 96.3 -177.1 -90.8 -120.7 11 A --- 133.0 56.2 139.6 178.0 -98.1 -95.5
Значение торсионных углов, их средние значения и отклонения представленны в таблице. Самый "деформированный" нуклеотид - это второй нуклеотид (А) на первой цепи.
Определение структуры водородных связей
Strand I Strand II Helix
1 (0.017) C:...2_:[..G]G-----C[..C]:..71_:C (0.004) |
2 (0.006) C:...3_:[..C]C-----G[..G]:..70_:C (0.006) |
3 (0.008) C:...4_:[..G]G-----C[..C]:..69_:C (0.005) |
4 (0.004) C:...5_:[..G]G-----C[..C]:..68_:C (0.014) |
5 (0.009) C:...6_:[..G]G-----C[..C]:..67_:C (0.009) |
6 (0.006) C:...7_:[..G]Gx----C[..C]:..66_:C (0.003) |
7 (0.005) C:..49_:[..G]G-----C[..C]:..65_:C (0.006) |
8 (0.009) C:..50_:[..U]U-*---G[..G]:..64_:C (0.007) |
9 (0.006) C:..51_:[..C]C-----G[..G]:..63_:C (0.006) |
10 (0.015) C:..52_:[..G]G-----C[..C]:..62_:C (0.002) |
11 (0.009) C:..53_:[..G]G----xC[..C]:..61_:C (0.010) |
12 (0.013) C:..54_:[5MU]u-**-xA[..A]:..58_:C (0.005) |
13 (0.043) C:..55_:[PSU]Px**+xG[..G]:..18_:C (0.007) x
14 (0.005) C:..35_:[..A]Ax*---U[..U]:..33_:C (0.005) |
15 (0.005) C:..38_:[..A]A-*---c[OMC]:..32_:C (0.009) |
16 (0.005) C:..39_:[..C]C-----G[..G]:..31_:C (0.013) |
17 (0.006) C:..40_:[..C]C-----G[..G]:..30_:C (0.008) |
18 (0.004) C:..41_:[..C]C-----G[..G]:..29_:C (0.006) |
19 (0.007) C:..42_:[..G]G-----C[..C]:..28_:C (0.005) |
20 (0.004) C:..43_:[..A]A-----U[..U]:..27_:C (0.006) |
21 (0.012) C:..44_:[..A]Ax*---G[..G]:..26_:C (0.010) |
22 (0.015) C:..10_:[..G]G-----C[..C]:..25_:C (0.007) |
23 (0.005) C:..11_:[..A]A-----U[..U]:..24_:C (0.008) |
24 (0.007) C:..12_:[..G]G-----C[..C]:..23_:C (0.005) |
25 (0.009) C:..13_:[..C]C----xG[..G]:..22_:C (0.004) |
26 (0.008) C:..14_:[..A]A-**-xu[4SU]:...8_:C (0.006) |
27 (0.007) C:..15_:[..G]Gx**+xC[..C]:..48_:C (0.006) x
28 (0.019) C:..19_:[..G]Gx---xC[..C]:..56_:C (0.004) +
В соответствии с полученными данными можно определить стебли:
Неканонические пары оснований в структуре:
Дополнительные водородные связи в тРНК:
Стэкинг-взаимодействия в РНК
Overlap area in Angstrom^2 between polygons defined by atoms on successive
bases. Polygons projected in the mean plane of the designed base-pair step.
Values in parentheses measure the overlap of base ring atoms only. Those
outside parentheses include exocyclic atoms on the ring. Intra- and
inter-strand overlap is designated according to the following diagram:
i2 3' 5' j2
/|\ |
| |
Strand I | | II
| |
| |
| \|/
i1 5' 3' j1
step i1-i2 i1-j2 j1-i2 j1-j2 sum
1 GC/GC 3.12( 1.31) 0.00( 0.00) 0.00( 0.00) 3.93( 1.84) 7.05( 3.15)
2 CG/CG 0.41( 0.03) 0.00( 0.00) 3.52( 0.90) 0.00( 0.00) 3.93( 0.93)
3 GG/CC 2.17( 0.75) 0.00( 0.00) 0.82( 0.00) 0.00( 0.00) 2.99( 0.75)
4 GG/CC 2.82( 1.24) 0.00( 0.00) 1.22( 0.00) 0.00( 0.00) 4.03( 1.24)
5 GG/CC 1.92( 0.53) 0.00( 0.00) 0.27( 0.00) 0.04( 0.00) 2.22( 0.53)
6 GG/CC 2.54( 1.05) 0.00( 0.00) 0.02( 0.00) 0.06( 0.00) 2.62( 1.05)
7 GU/GC 5.43( 2.45) 0.00( 0.00) 0.00( 0.00) 7.29( 3.77) 12.72( 6.22)
8 UC/GG 0.61( 0.00) 0.00( 0.00) 0.25( 0.00) 1.84( 0.52) 2.70( 0.52)
9 CG/CG 1.80( 0.70) 0.00( 0.00) 2.36( 0.35) 0.00( 0.00) 4.16( 1.05)
10 GG/CC 2.73( 1.23) 0.00( 0.00) 0.75( 0.00) 0.00( 0.00) 3.48( 1.23)
11 Gu/AC 8.26( 3.56) 0.00( 0.00) 0.00( 0.00) 3.48( 1.21) 11.74( 4.77)
12 uP/GA 5.33( 0.21) 0.00( 0.00) 0.00( 0.00) 4.31( 1.89) 9.65( 2.10)
13 PA/UG 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
14 AA/cU 6.15( 4.41) 0.00( 0.00) 0.00( 0.00) 3.80( 1.70) 9.95( 6.11)
15 AC/Gc 1.01( 0.76) 0.00( 0.00) 0.00( 0.00) 7.05( 2.84) 8.07( 3.60)
16 CC/GG 0.00( 0.00) 0.00( 0.00) 1.23( 0.00) 3.05( 1.52) 4.28( 1.52)
17 CC/GG 0.00( 0.00) 0.00( 0.00) 0.45( 0.00) 3.61( 2.12) 4.07( 2.12)
18 CG/CG 0.00( 0.00) 0.00( 0.00) 4.35( 1.63) 0.00( 0.00) 4.35( 1.63)
19 GA/UC 3.42( 1.99) 0.00( 0.00) 0.00( 0.00) 0.59( 0.00) 4.02( 1.99)
20 AA/GU 1.54( 0.67) 0.00( 0.00) 0.00( 0.00) 5.08( 2.37) 6.62( 3.03)
21 AG/CG 0.00( 0.00) 0.00( 0.00) 3.10( 1.09) 0.00( 0.00) 3.10( 1.09)
22 GA/UC 1.12( 0.00) 0.00( 0.00) 0.63( 0.00) 0.00( 0.00) 1.75( 0.00)
23 AG/CU 1.15( 1.15) 0.00( 0.00) 0.46( 0.00) 0.53( 0.01) 2.13( 1.16)
24 GC/GC 6.25( 2.34) 0.00( 0.00) 0.00( 0.00) 4.40( 1.88) 10.65( 4.22)
25 CA/uG 0.00( 0.00) 0.37( 0.00) 5.92( 4.78) 0.00( 0.00) 6.28( 4.78)
26 AG/Cu 2.78( 0.79) 0.00( 0.00) 0.01( 0.00) 0.00( 0.00) 2.79( 0.79)
27 GG/CC 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
28 GG/CC 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
29 GC/GC 3.37( 1.39) 0.00( 0.00) 0.00( 0.00) 3.81( 1.75) 7.17( 3.14)
30 CG/CG 0.13( 0.01) 0.00( 0.00) 3.86( 1.13) 0.00( 0.00) 3.99( 1.14)
31 GG/CC 2.78( 1.29) 0.00( 0.00) 0.54( 0.00) 0.07( 0.00) 3.39( 1.29)
32 GG/CC 3.06( 1.50) 0.00( 0.00) 0.95( 0.00) 0.00( 0.00) 4.01( 1.50)
33 GG/CC 1.53( 0.31) 0.00( 0.00) 0.46( 0.00) 0.07( 0.00) 2.07( 0.31)
34 GG/CC 2.28( 0.65) 0.00( 0.00) 0.00( 0.00) 0.53( 0.11) 2.82( 0.75)
35 GU/GC 5.15( 2.22) 0.00( 0.00) 0.00( 0.00) 6.80( 3.32) 11.95( 5.55)
36 UC/GG 1.31( 0.04) 0.00( 0.00) 0.06( 0.00) 1.78( 0.45) 3.15( 0.49)
37 CG/CG 1.57( 0.33) 0.00( 0.00) 1.93( 0.13) 0.01( 0.00) 3.51( 0.46)
38 GG/CC 3.00( 1.47) 0.00( 0.00) 0.46( 0.00) 0.00( 0.00) 3.46( 1.47)
39 Gu/AC 8.28( 3.28) 0.00( 0.00) 0.00( 0.00) 3.67( 1.35) 11.95( 4.63)
40 uP/GA 4.71( 0.24) 0.00( 0.00) 0.00( 0.00) 3.22( 1.13) 7.93( 1.37)
41 PA/UG 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)
42 AA/cU 5.09( 3.72) 0.00( 0.00) 0.00( 0.00) 5.93( 3.05) 11.02( 6.76)
43 AC/Gc 2.39( 1.52) 0.00( 0.00) 0.00( 0.00) 7.46( 4.50) 9.85( 6.02)
44 CC/GG 0.00( 0.00) 0.00( 0.00) 0.95( 0.00) 3.13( 1.61) 4.08( 1.61)
45 CC/GG 0.00( 0.00) 0.00( 0.00) 0.43( 0.00) 3.97( 2.57) 4.41( 2.57)
46 CG/CG 0.00( 0.00) 0.00( 0.00) 4.54( 1.81) 0.00( 0.00) 4.54( 1.81)
47 GA/UC 2.98( 1.48) 0.00( 0.00) 0.00( 0.00) 0.51( 0.00) 3.49( 1.48)
48 AA/GU 1.18( 0.46) 0.00( 0.00) 0.00( 0.00) 5.10( 2.41) 6.29( 2.87)
49 AG/CG 0.00( 0.00) 0.00( 0.00) 2.31( 0.65) 0.00( 0.00) 2.31( 0.65)
50 GA/UC 1.06( 0.00) 0.00( 0.00) 0.82( 0.00) 0.00( 0.00) 1.88( 0.00)
51 AG/CU 1.11( 1.08) 0.00( 0.00) 0.54( 0.00) 0.32( 0.00) 1.97( 1.08)
52 GC/GC 5.27( 1.38) 0.00( 0.00) 0.00( 0.00) 3.79( 1.54) 9.06( 2.93)
53 CA/uG 0.00( 0.00) 0.52( 0.00) 5.68( 4.64) 0.00( 0.00) 6.21( 4.64)
54 AG/Cu 2.42( 0.63) 0.00( 0.00) 0.01( 0.00) 0.00( 0.00) 2.43( 0.63)
55 GC/AC 2.66( 0.49) 0.59( 0.00) 0.00( 0.00) 0.00( 0.00) 3.25( 0.49)
56 CG/CA 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00) 0.00( 0.00)