Optimal pairwise alignment
Last updated: 26-04-2017.
Files to download
Task 1. Global and local alignment comparison
Alignments for the first task were received using Emboss programs 'needle' and 'water'. Main information about them is presented in Table 1, alignments themselves are presented in Figures 1 and 2. Global alignment length is 658, local one is 612. So, local alignment is shorter than global one. Main differences betwixt global and local alignments are in algorithms themselves which are discussed in tasks 4 and 5. Global alignment suggests that observed sequences are homologous along the entire length, local alignment admits that proteins may contain non-homologous plots just as homologous ones. Using figures one and two it is not hard to notice, that global and local alignments of HS71L_MOUSE (P16627) and DNAK_METBF (Q465Y6) are almost absolutely the same: local alignment is a global one without 1..10 positions.


| Sequence name | AC | Alignment type | Sequence length | Alignment length | Amount of conservative residues | Percent of conservative residues | Amount of functional-conservative residues | Percent of functional-conservative residues | Amount of gaps | Percent of gaps | Amount indels |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Homologous sequences | |||||||||||
| HS71L_MOUSE/1-641 | P16627 | GLOBAL | 641 | 658 | 290 | 44,1 % | 406 | 61,7 % | 13 | 2,0 % | 5 |
| DNAK_METBF/1-620 | Q465Y6 | GLOBAL | 620 | 658 | 290 | 44,1 % | 406 | 61,7 % | 38 | 5,8 % | 8 |
| HS71L_MOUSE/1-641 | P16627 | LOCAL | 606 | 612 | 282 | 46,1 % | 396 | 64,7 % | 6 | 1,0 % | 2 |
| DNAK_METBF/1-620 | Q465Y6 | LOCAL | 574 | 612 | 282 | 46,1 % | 396 | 64,7 % | 38 | 6,2 % | 8 |
| Non-homologous sequences | |||||||||||
| 1 | |||||||||||
| MALE_SALTY | P19576 | LOCAL | 312 | 379 | 77 | 20,3 % | 127 | 33,5 % | 67 | 17,7 % | 16 |
| PLED_CAUCN | B8GZM2 | LOCAL | 316 | 379 | 77 | 20,3 % | 127 | 33,5 % | 63 | 16,6 % | 9 |
| 2 | |||||||||||
| MALE_SALTY | P19576 | LOCAL | 42 | 42 | 11 | 26,2 % | 21 | 50,0 % | 0 | 0,0 % | 0 |
| FMM1_NEIGO | P02974 | LOCAL | 35 | 42 | 11 | 26,2 % | 21 | 50,0 % | 7 | 1,7 % | 1 |
| 3 | |||||||||||
| MALE_SALTY | P19576 | LOCAL | 98 | 118 | 25 | 21,2 % | 39 | 33,1 % | 20 | 17,0 % | 3 |
| Y2262_MYCBO | P65689 | LOCAL | 92 | 118 | 25 | 21,2 % | 39 | 33,1 % | 26 | 22,0 % | 5 |
| 4 | |||||||||||
| MALE_SALTY | P19576 | LOCAL | 94 | 147 | 35 | 23,8 % | 54 | 36,7 % | 53 | 36,1 % | 6 |
| HETS_PODAS | Q03689 | LOCAL | 140 | 147 | 35 | 23,8 % | 54 | 36,7 % | 7 | 4,8 % | 3 |
| 5 | |||||||||||
| MALE_SALTY | P19576 | LOCAL | 331 | 386 | 79 | 20,5 % | 131 | 44,0 % | 55 | 14,3 % | 11 |
| A0A126V644_9RHOB | A0A126V644 | LOCAL | 312 | 386 | 79 | 20,5 % | 131 | 44,0 % | 74 | 19,2 % | 12 |
Table 1. Some information about observed alignments.
| Attribute | Global | Local |
|---|---|---|
| Score matrix | BLOSUM62 | BLOSUM62 |
| Indel opening penalty | 10 | 10 |
| Indel extension penalty | 0,5 | 0,5 |
| End gap penalty applying | No | - |
| End gap opening penalty | 10 | - |
| End gap extending penalty | 0,5 | - |
Table 2. Alignment parameters.
Task 2. Local alignment of homologous and non-homologous sequences comparison
According to Table 1, Figures 2 and 3, local alignments of non-homologous sequences are most likely shorter than alignments of homologous sequences. Also, they have less percent of conservative and functional-conservative residues. Talking about biological sense, local alignment of non-homologous sequences seems senseless and not informative at all: local alignment algorithm is suitable for aligning homologous domain proteins.
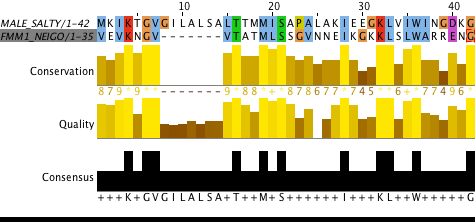
Task 3. Multiple, local and global alignments comparison
HS71L_MOUSE (P16627) and DNAK_METBF (Q465Y6) sequences were chosen for this task. All alignments are seem similar but there is a couple of differences betwixt them, for example: in multiple alignment VAL(80) stands against TYR(109), in local and global alignments VAL(80) stands against VAL(107); in multiple alignment THR(81) stands against LYS(110), in local and global alignments THR(81) stands against SER(108); in multiple alignment LEU(82) stands against GLY(111), in local and global alignments LEU(82) stands against TYR(109). Most of differences are located in the beginning and the end of sequences.

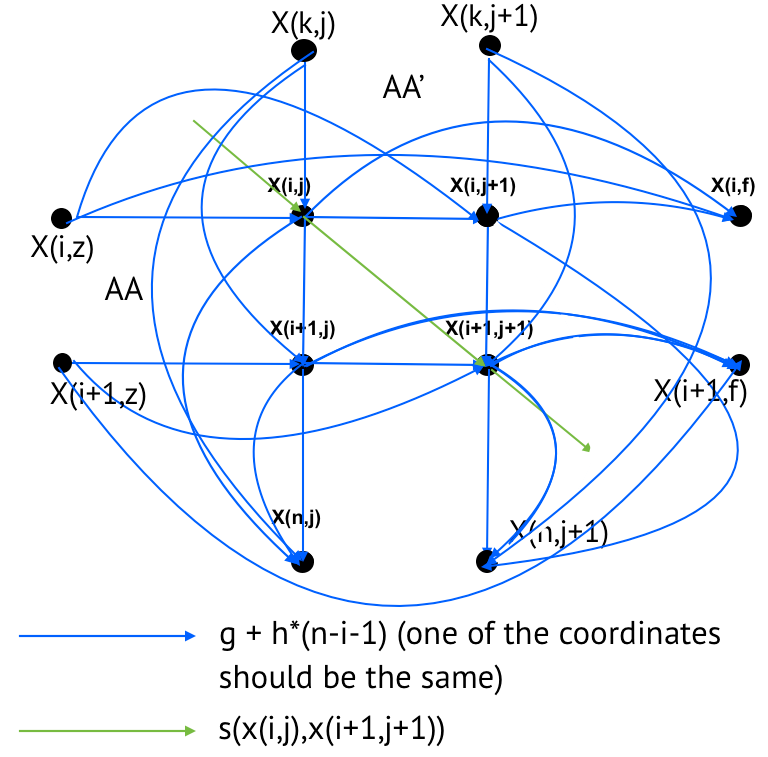
Task 4. Global alignment with affine penalties graph
Graph of global alignment with affine penalties is presented in Figure 5.
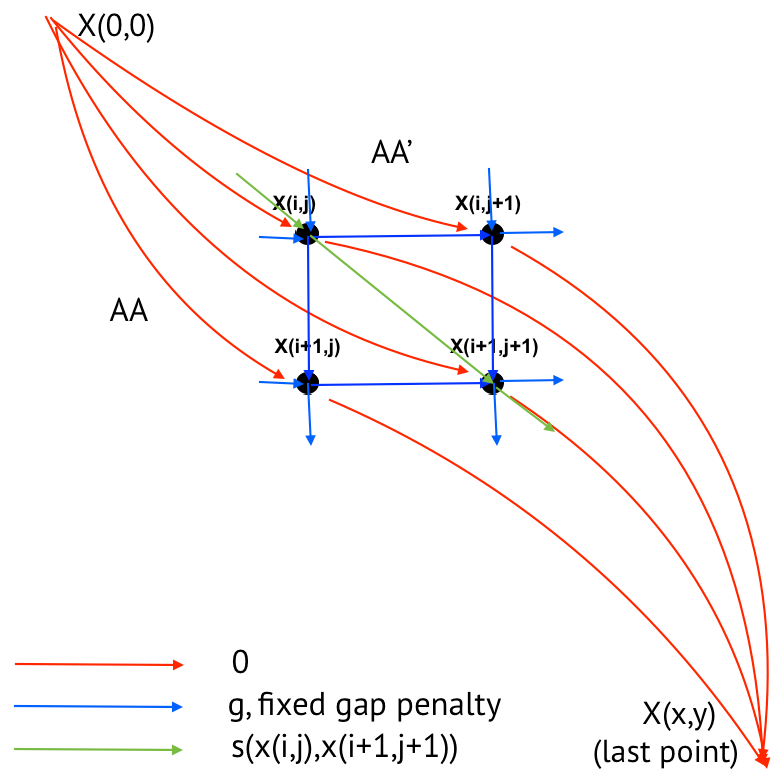
Task 5. Local alignment with linear penalties graph
Graph of local alignment with linear penalties is presented in Figure 6.
Task 6. "Scores of friendliness"
Seat scheme is presented in Figure 7 and Insertion 1. The probability of meeting a male q(M) is 0.53, female q(F) is 0.47. Amount of pairs is 6*5 = 30. If the audience seated casually and independently of each other, then there is q(M)*q(M)*30 = 8.4 of MM pairs, 6.6 FF pairs, 7.4 pairs of FM and MF. Actual shares of pairs is: p(MM) = 0.27, p(FF) = 0.2 p(FM) = 0.27. The inclination of men to sit next to each other L(MM) is p(MM)/q(M)*q(M), similarly for other pairs. For example, -100*log_2(L(MM)) is called male-male score of friendliness. Table of scores of friendliness has number 3.
MFMMMF MFMFFM FFMMFF FFMMMM MFFMMF MFFMMF
Insertion 1. Seat scheme.

| M | F | Sum | |
|---|---|---|---|
| M | 496 | 478 | 974 |
| F | 478 | 504 | 982 |
| Sum | 974 | 982 |
Table 3. Scores of friendliness