Proteins and macromolecular complexes interior
Last updated: 21-02-2017.
Information about the script which defines hydrophobic core
The original script from CluD[0] (click here to download) is a little bit irrational because it defines atoms row by row using ‘define’ command in every line. So, using specially created file ‘test.txt’ (with atoms names) and simple python program (insertion 1), more rational script was created (click here to download).
import urllib2
indata = urllib2.urlopen("http://kodomo.fbb.msu.ru/~simon/term2/test.txt")
x = []
for i in indata:
x.append(i.split()[0])
out = str('define core2 core2,GLU3:A.CB')
for i in range(1, len(x)):
out += (' or ' + x[i])
print outInsertion 1. Python program.
Download scripts
[download script №1] [download script №2] [download script №3] [download script №4]
Information about hydrophobic core
Hydrophobic core contains 734 atoms, which is 21.89% of all atoms described in pdb file (protein + water + ligand) or 25.47% of protein atoms. This core occupies almost the entire interior of protein, so I suggest that it helps to keep the protein's structure. Fig. 1 proves this conjecture and shows that protein has a special shape for ligand binding.
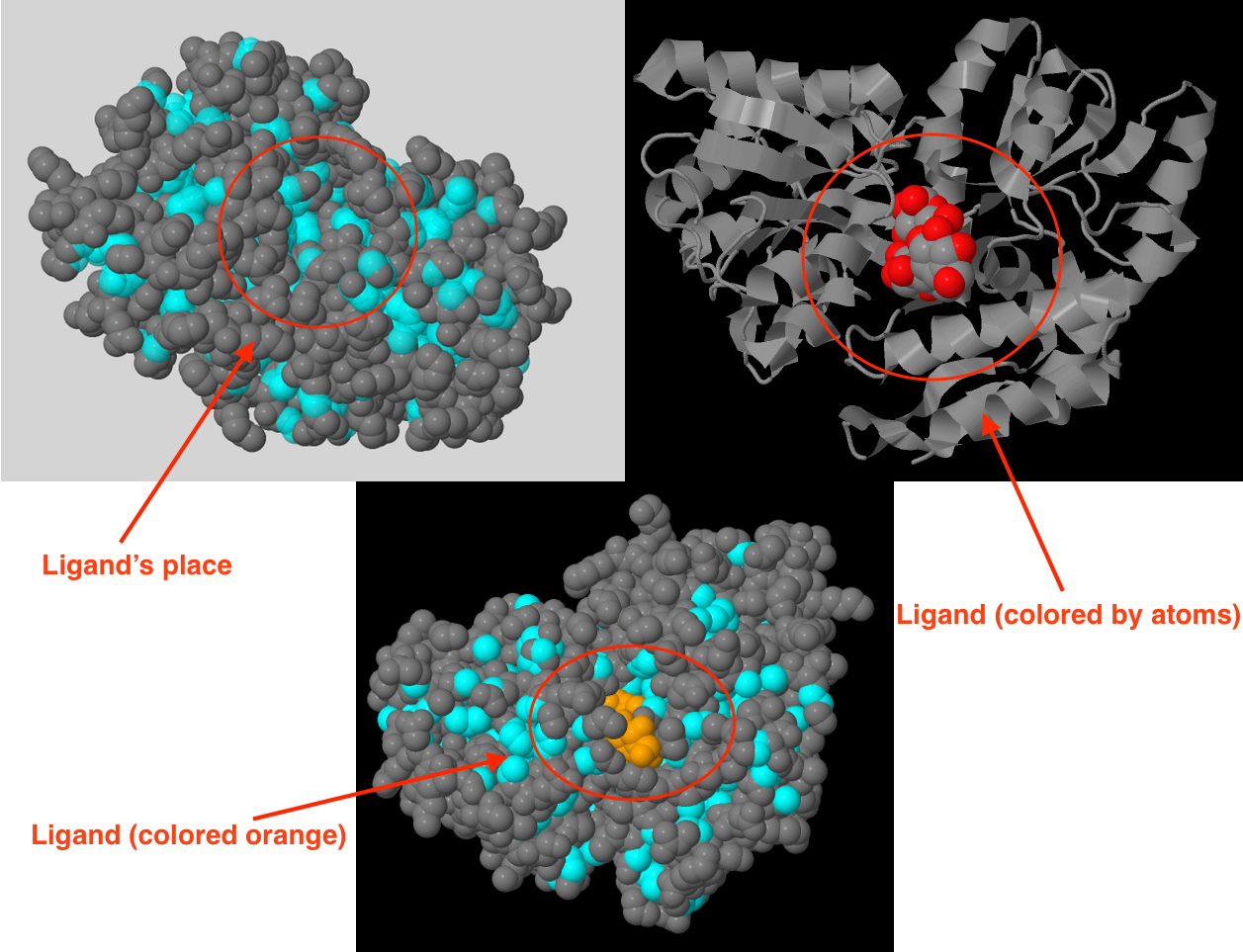
Atoms packaging density in hydrophobic core
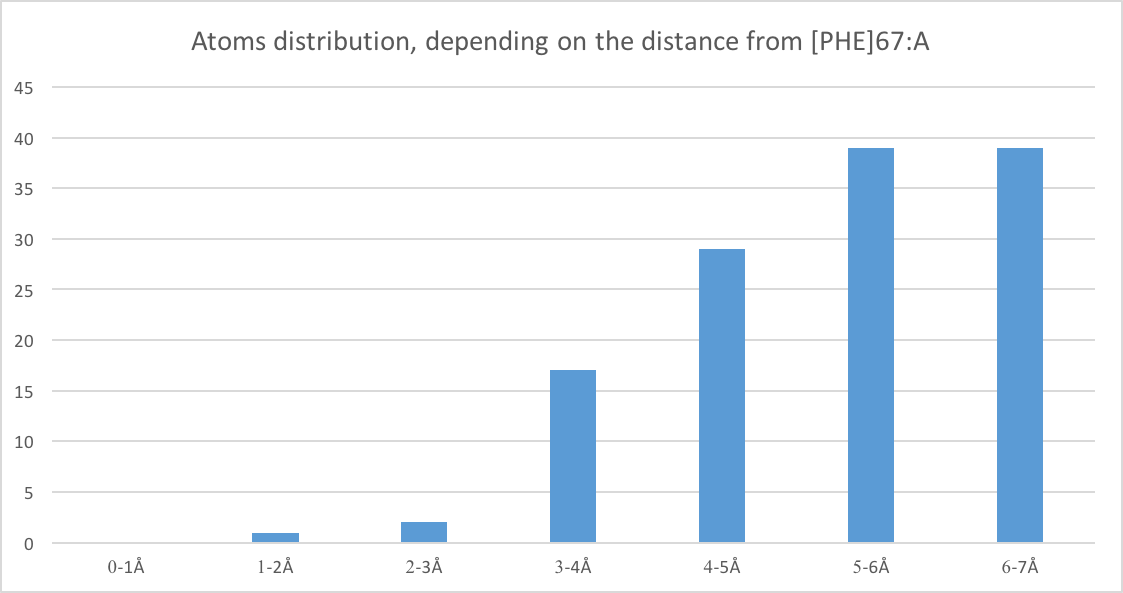
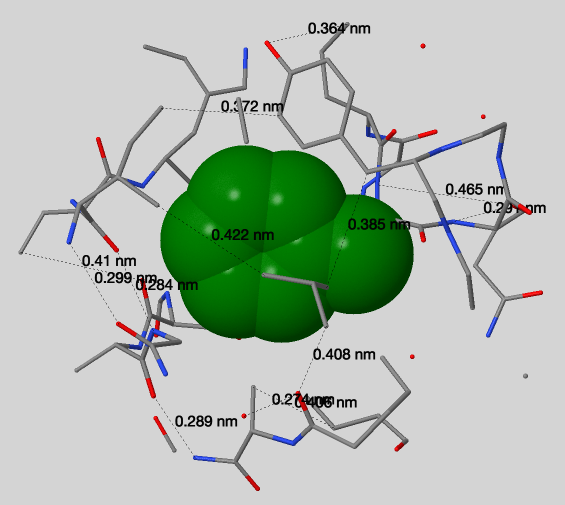
Fig. 2 represents atoms distribution, depending on the distance from [PHE]67:A. This histogram is based on the results obtained using script №2. Also, applying this script it can be seen that atoms at a distance of six Å from the [PHE]67:A are almost completely cover this residue (but it is still possible to catch sight of it). Using J(S)Mol measures (Fig. 3) we can conclude that typical distance between two not bonded covalently atoms is 3.68Å (median from Table 1) which is not enough for water to pass through (average should be at least 1.4 (oxygen Van der Vaals radius) + 1.4 + 1.4*2 = 5.6Å).
| № of measure | Distance, Å |
|---|---|
| 1 | 2.74 |
| 2 | 4.08 |
| 3 | 4.22 |
| 4 | 2.99 |
| 5 | 4.10 |
| 6 | 3.29 |
| 7 | 3.64 |
| 8 | 4.65 |
| 9 | 2.91 |
| 10 | 4.06 |
| 11 | 2.89 |
| 12 | 2.84 |
| 13 | 3.85 |
| 14 | 3.72 |
Table 1. Distances between not bonded covalently atoms around [PHE]67:A.
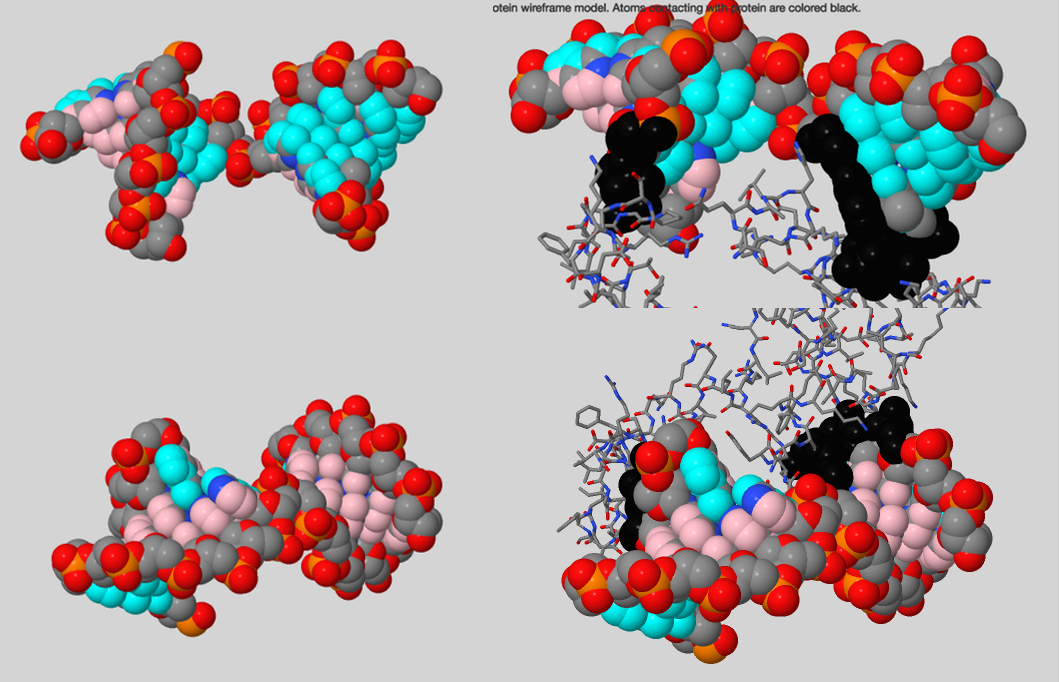
Protein-DNA complex
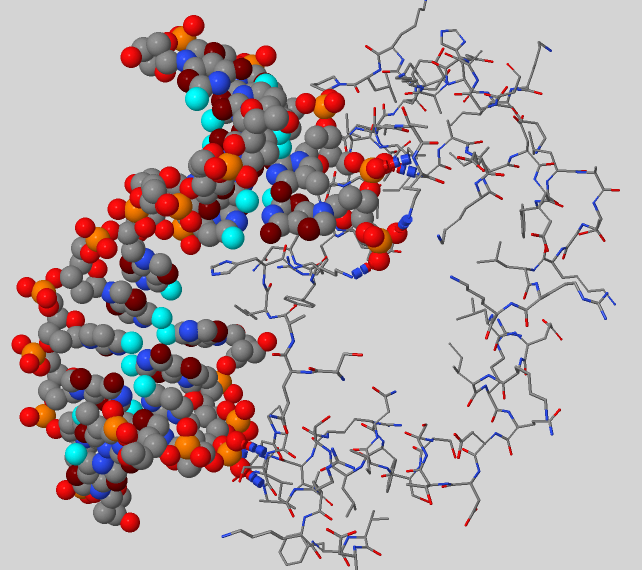
Subject protein has an index 5J0O and occurs to be the DNA polymerase beta and performs base excision repair required for DNA maintenance, replication, recombination, and damaging factors resistance [1]. But proteins interacting with DNA can perform a bunch of other functions:
DNA compartments and donors and acceptors that can be involved into hydrogen bonds with proteins
Using script №4 it was found out that protein interacts with DNA backbone, its major groove and less with minor groove (Fig. 4). But hydrogen bonds were observed only between protein and DNA backbone (Fig.5). Noticeable observation: using information about donors and acceptors in major groove we can establish DNA sequence.
Reaction mechanism
Reaction shown in the video is a typical bimolecular nucleophilic substitution. Reaction can be generally explained as Nuc: + R-LG → R-Nuc + LG: ("an electron rich nucleophile selectively bonds with or attacks the positive or partially positive charge of an atom or a group of atoms to replace a leaving group; the positive or partially positive atom is referred to as an electrophile. The whole molecular entity of which the electrophile and the leaving group are part is usually called the substrate") (Fig. 6)[2]. The process proceeds in the video is a dephosphorylation of paraoxon, irreversible acetylcholinesterase inhibitor that used as very powerful insecticide and chemical weapon[3].
References
[0] CluD service. [1] DNA polymerase beta, Wikipedia. [2] Nucleophilic substitution, Wikipedia. [3] Paraoxon, Wikipedia.