Analysis of major genome rearrangements
Last updated: 19-11-2017.
Files to download
[blocks.gbi] [pangenome.bi] [pangenome.info] [excel file with calculations from task 2]
Task 1. Chosen genomes
- Salmonella enterica subsp. enterica serovar Enteritidis str. EC20121176 (CP007270.2)
- Salmonella enterica subsp. enterica serovar Montevideo str. 507440-20 (CP007530.1)
- Salmonella enterica subsp. enterica serovar Gallinarum/pullorum str. RKS5078 (CP003047.1)
Task 2. Identity % and coverage of genomes by homologous sites
Total length of homologous sites (= 4068960) were calculated using blocks.gbi file generated by NPG-explorer (you can download file above in chapter 'files to download'). Actually, it wasn't essential because pangenome.info file contained this information, too. Summary information about coverage of genomes is presented in Table 1. Information about identity % was obtained from pangenome.info from chapter describing 'stem blocks'. Value of identity % is 0.984486.
| Genome length | Coverage % | |
| Enteritidis | 4685848 | 86,8 % |
| Montevideo | 4694375 | 86,7 % |
| Gallinarum | 4637962 | 87,7 % |
Table 1. Coverage of genomes by homologous sites.
Task 3. Genome rearrangements
Deletions and insertions
Example for this task was found in file pangenome.bi. Block h2x3003 was chosen. It contains gene of endonuclease (CP007270.2). I suggest that it is more possibly insertion than deletion due to that fact that observed CDS is located in region associated with phages:
/region_name="holin_lambda" /note="phage holin, lambda family; TIGR01594"
Salmonella enterica subsp. enterica serovar Enteritidis str. EC20121176 and Salmonella enterica subsp. enterica serovar Gallinarum/pullorum str. RKS5078 (CP003047.1) have this endonuclease in their genomes, so it is possible to suggest that they are more related to each other than Salmonella enterica subsp. enterica serovar Montevideo str. 507440-20 to them. Or they were infected by the same phage.
Inversions
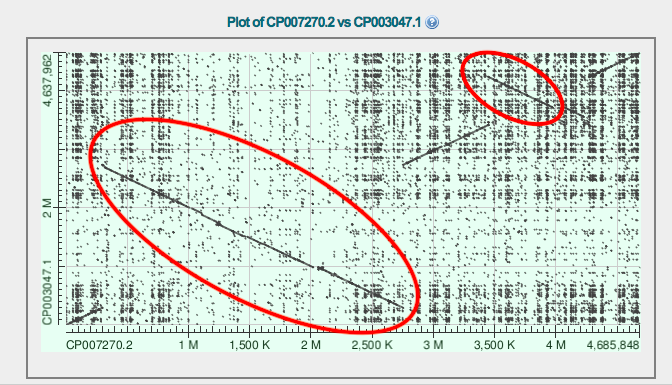
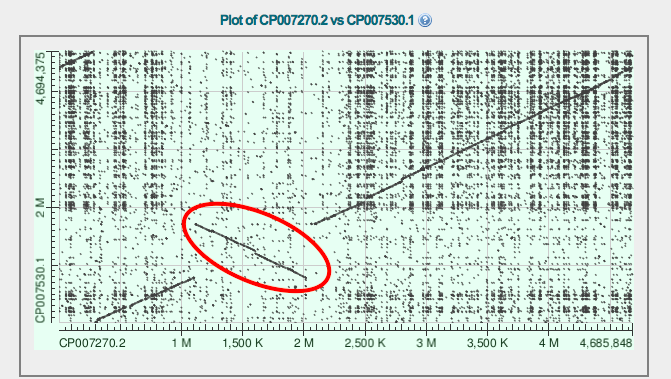
For this task I used blast2seq: Salmonella enterica subsp. enterica serovar Enteritidis str. EC20121176 against Salmonella enterica subsp. enterica serovar Gallinarum/pullorum str. RKS5078 (Expires on 11-21 01:11 am) and Salmonella enterica subsp. enterica serovar Enteritidis str. EC20121176 against Salmonella enterica subsp. enterica serovar Montevideo str. 507440-20 (Expires on 11-21 01:10 am) Examples of inversions are marked red in Fig.1 and 2.