DNA-protein complexes
Last updated: 01-10-2017.
Files to download
Task 1. Prediction of the secondary structure of observed tRNA
Outcome of einverted program was empty with standard parameters, so custom ones were used. They are presented in the insertion below. Outcome itself is presented in Fig. 1. RNAfold outcome was very close to the reality from the very first attempt with default parameters. Figure 2 displays graphical image of suggested structure and probability distribution of nucleotide pair formation. Results of comparison 'find_pair', 'einverted' and 'RNAfold' programs are presented in Table 1.
Gap penalty [12]: 5 Minimum score threshold [50]: 20 Match score [3]: 5 Mismatch score [-4]: -1

| Part of the structure | Real positions according to find_pair | Predicted amount of pairs by einverted | Predicted amount of pairs by the Zuker algorithm |
|---|---|---|---|
| Acceptor stem | 5'-1-7-3' 5'-66-72-3' 7 pairs | 6 | 7 |
| T-stem | 5'-49-53-3' 5'-61-65-3' 5 pairs | 0 | 5 |
| D-stem | 5'-10-13-3' 5'-22-25-3' 4 pairs | 0 | 4 |
| Anticodon stem | 5'-36-44-3' 5'-26-33-3' 8 pairs | 5 | 6 A-G and P-A pairs are missed |
| Total amount of canonical pairs | 20 | 11 | 18 |
Table 1. Comparison of programs dealing with tRNA secondary structure.
Task 2. DNA-protein contacts in observed structure (1cf7)
Part 1. Definition of the set of atoms
According to the task the following sets were defined:
- set1: oxygen atoms of 2'-deoxyribose
- set2: oxygen atoms of phosphoric acid
- set3: nitrigen atoms of nucleotides
The results of the task are available to view in JMol applet (click on the applet and then 'Load Script №1 button). Use 'Resume' button to resume.
Part 2. DNA-protein contacts
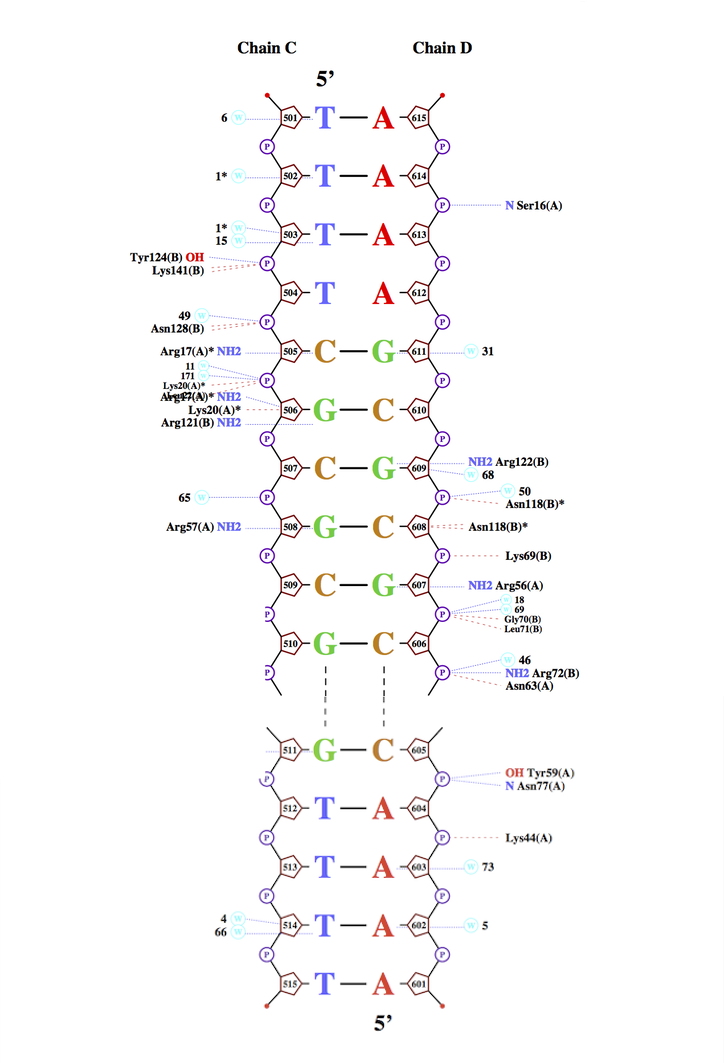
Сomprehensive information about studied bonds is presented in JMol applet (Script №2). DNA's interacting atoms are colored red, protein's interacting atoms are colored blue. Brief summary about amount of each interaction type is presented in Table 2.
| Protein contacts | Polar | Non-polar | Total amount |
|---|---|---|---|
| with the residues of 2'-deoxyribose | 8 | 34 | 42 |
| with residues of phosphoric acid | 15 | 11 | 26 |
| with nucleotide residues of major groove | 9 | 2 | 36 |
| with nucleotide residues of minor groove | 2 | 9 | 11 |
Table 2. Information about DNA-protein contacts.
Part 3. Nucplot
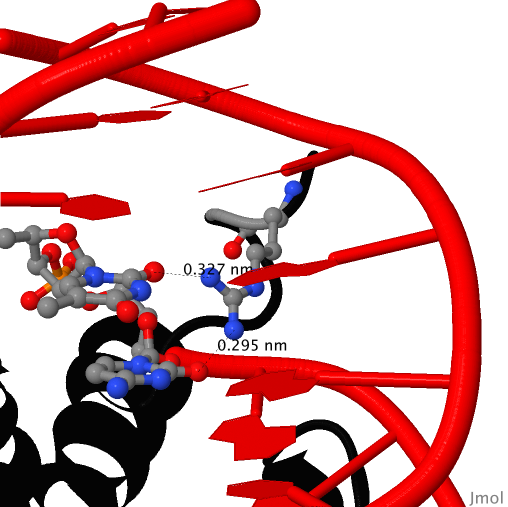
According to the nucplot outcome (Fig. 3), amino acid residue with the largest number of contacts (2) is Arg 17. This residue is interacting with cytosine ([DC]505 in .pdb) and thymine ([DT]504 in .pdb). This interaction is presented in Fig. 4. As it seen in Fig. 3, Arg 17 is located in kind of binding site (in a cluster of amino acids interacting with DNA), so it is logical to assume that Arg 17 is the most important residue for DNA recognition considering the fact that Arg 17 interacts with two nucleotides.