Sanger sequencing
Last updated: 17-10-2017.
Files to download
[Archive containing .ab1 files from Task 1]
[.jvp project from Task 1] [.ab1 file from Task 2]
Task 1. Сhromatograms
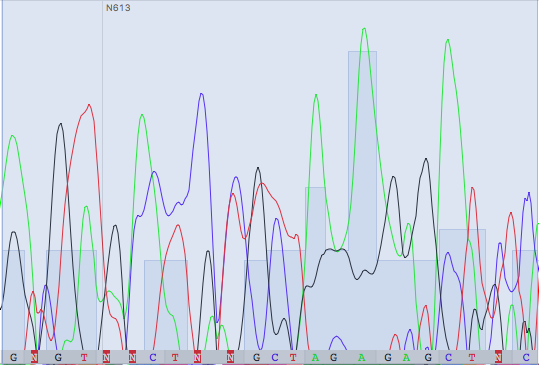
Unreadable parts were found and deleted: in the direct read 1-20 and 675-718, and in the reverse read 1-21 and 673-720 ( (-37) - (-18) and 636-683 by the numbering of a straight chain in alignment). It is strange that the lengths of the initial unreadable sections in the chains are approximately the same as the lengths of the final ones, although everything should vice versa, due to the use of "reverse complement", the beginning of the reverse read is turned out to be the end. Final alignment is presented in Fig.1. Table 1 explains some problems were solved during correcting of reads.

| Picture | Description |
|---|---|
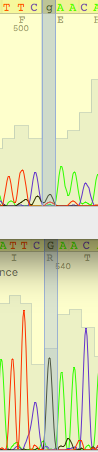 | The signal was extremely low and indistinguishable from noise. Reverse read clarified the situation |
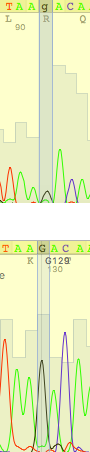 | Situation is the same as previous one (but problem was caused by reverse read) |
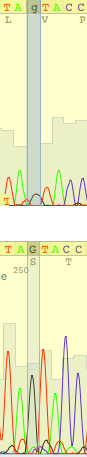 | Guanine caused the problem again! |
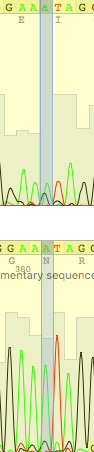 | This situation could be explained as polymorphism, but reverse read brings unambiguity |
Table 1. Main parameters of studied DNA forms.
Task 2. Unreadable part of chromatogram
For this task kamp3_18SIII_F_F03_WSBS-Seq-1-08-15.ab1 file was used. It's impossible to read this chromatogram due to blurred peaks, high noise level, peak offset and pollution of the sample. Part of file is presented in Fig. 2