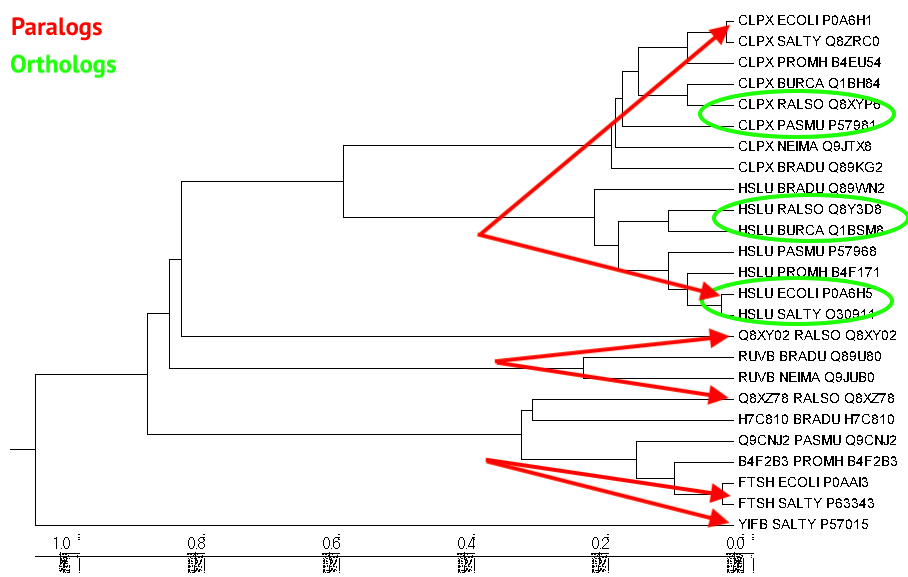
Orthologs and paralogs
Last updated: 15-03-2018.
Process of data preparation
The bacteria for this task were taken from previous task
The following steps were taken:
File containing genomes of all bacteria was created:
cat /P/y16/term4/Proteomes/BRADU.fasta >> united_genomes cat /P/y16/term4/Proteomes/BURCA.fasta >> united_genomes cat /P/y16/term4/Proteomes/RALSO.fasta >> united_genomes cat /P/y16/term4/Proteomes/NEIMA.fasta >> united_genomes cat /P/y16/term4/Proteomes/SALTY.fasta >> united_genomes cat /P/y16/term4/Proteomes/PASMU.fasta >> united_genomes cat /P/y16/term4/Proteomes/PROMH.fasta >> united_genomes cat /P/y16/term4/Proteomes/ECOLI.fasta >> united_genomes
Blast database was created:
makeblastdb -in united_genomes.fasta -dbtype prot
Blast search was performed:
blastp -query CLPX_ECOLI.fasta -db united_genomes.fasta -evalue 0.001 -outfmt 7
File for seqret was created:
echo united_genomes.fasta:Q8ZRC0 >> file.fasta echo united_genomes.fasta:B4EU54 >> file.fasta echo united_genomes.fasta:Q1BH84 >> file.fasta echo united_genomes.fasta:Q8XYP6 >> file.fasta echo united_genomes.fasta:P57981 >> file.fasta echo united_genomes.fasta:Q89KG2 >> file.fasta echo united_genomes.fasta:Q9JTX8 >> file.fasta echo united_genomes.fasta:Q8Y3D8| >> file.fasta echo united_genomes.fasta:Q8Y3D8 >> file.fasta echo united_genomes.fasta:B4F171 >> file.fasta echo united_genomes.fasta:P0A6H5 >> file.fasta echo united_genomes.fasta:P57968 >> file.fasta echo united_genomes.fasta:Q89WN2 >> file.fasta echo united_genomes.fasta:O30911 >> file.fasta echo united_genomes.fasta:Q1BSM8 >> file.fasta echo united_genomes.fasta:B4F2B3 >> file.fasta echo united_genomes.fasta:P0AAI3 >> file.fasta echo united_genomes.fasta:P63343 >> file.fasta echo united_genomes.fasta:Q89U80 >> file.fasta echo united_genomes.fasta:A0A0H2XMS5 >> file.fasta echo united_genomes.fasta:H7C810 >> file.fasta echo united_genomes.fasta:Q8XZ78 >> file.fasta echo united_genomes.fasta:Q9CNJ2 >> file.fasta echo united_genomes.fasta:|Q9JUB0 >> file.fasta echo united_genomes.fasta:Q9JUB0 >> file.fasta echo united_genomes.fasta:Q8XY02 >> file.fasta echo united_genomes.fasta:P57015 >> file.fasta
Fasta file containing sequences was generated:
seqret -seq @file.fasta -out seqs.fasta
Muscle alignment:
muscle -in seqs.fasta -out align.fasta
MEGA
Tree of found homologous proteins was created using UPGMA method by MEGA7 program. Tree is presented in Fig. 1. You can also observe some examples of orthologs and paralogs in this figure.