Задание 10. Гомологичное моделирование комплекса белка с лигандом
Цель данного занятия ознакомится с возможностями гомологичного моделирования комплекса белка с лигандом. В этом занятии мы будем пользоваться пакетом Modeller. Это программное обеспечение распространяется бесплатно для академических пользователей.
Программе MODELLER на вход были поданы: 1. файл pdb со структурой-образцом 1lmp.pdb 2. выравнирание 1lmp и исследуемого белка LYS_BOMMO из тутового шелкопряда.
In [1]:
import sys
sys.path.append('/usr/lib/modeller9v7/modlib/')
sys.path.append('/usr/lib/modeller9v7/lib/x86_64-intel8/python2.5/')
import modeller
import _modeller
import modeller.automodel
In [2]:
env=modeller.environ()
env.io.hetatm=True
MODELLER 9v7, 2009/06/12, r6923
PROTEIN STRUCTURE MODELLING BY SATISFACTION OF SPATIAL RESTRAINTS
Copyright(c) 1989-2009 Andrej Sali
All Rights Reserved
Written by A. Sali
with help from
B. Webb, M.S. Madhusudhan, M-Y. Shen, M.A. Marti-Renom,
N. Eswar, F. Alber, M. Topf, B. Oliva, A. Fiser,
R. Sanchez, B. Yerkovich, A. Badretdinov,
F. Melo, J.P. Overington, E. Feyfant
University of California, San Francisco, USA
Rockefeller University, New York, USA
Harvard University, Cambridge, USA
Imperial Cancer Research Fund, London, UK
Birkbeck College, University of London, London, UK
Kind, OS, HostName, Kernel, Processor: 4, Linux kodomo.fbb.msu.ru 3.8-2-amd64 x86_64
Date and time of compilation : 2009/06/12 12:23:44
MODELLER executable type : x86_64-intel8
Job starting time (YY/MM/DD HH:MM:SS): 2017/05/24 14:48:16
In [3]:
! wget http://www.pdb.org/pdb/files/1lmp.pdb
--2017-05-24 14:48:19-- http://www.pdb.org/pdb/files/1lmp.pdb
Resolving www.pdb.org... 132.249.231.77
Connecting to www.pdb.org|132.249.231.77|:80... connected.
HTTP request sent, awaiting response... 301 Moved Permanently
Location: http://www.rcsb.org/pdb/files/1lmp.pdb [following]
--2017-05-24 14:48:20-- http://www.rcsb.org/pdb/files/1lmp.pdb
Resolving www.rcsb.org... 132.249.231.10
Connecting to www.rcsb.org|132.249.231.10|:80... connected.
HTTP request sent, awaiting response... 301 Moved Permanently
Location: http://files.rcsb.org/view/1lmp.pdb [following]
--2017-05-24 14:48:20-- http://files.rcsb.org/view/1lmp.pdb
Resolving files.rcsb.org... 132.249.213.140
Connecting to files.rcsb.org|132.249.213.140|:80... connected.
HTTP request sent, awaiting response... 200 OK
Length: unspecified [text/plain]
Saving to: `1lmp.pdb'
[ <=> ] 131,301 216K/s in 0.6s
2017-05-24 14:48:21 (216 KB/s) - `1lmp.pdb' saved [131301]
In [4]:
! wget http://www.uniprot.org/uniprot/LYS_BOMMO.fasta
--2017-05-24 14:48:22-- http://www.uniprot.org/uniprot/LYS_BOMMO.fasta Resolving www.uniprot.org... 128.175.240.211, 193.62.193.81 Connecting to www.uniprot.org|128.175.240.211|:80... connected. HTTP request sent, awaiting response... 302 Found Location: /uniprot/P48816.fasta [following] --2017-05-24 14:48:23-- http://www.uniprot.org/uniprot/P48816.fasta Reusing existing connection to www.uniprot.org:80. HTTP request sent, awaiting response... 200 OK Length: 195 [text/plain] Saving to: `P48816.fasta' 100%[======================================>] 195 --.-K/s in 0s 2017-05-24 14:48:23 (17.1 MB/s) - `P48816.fasta' saved [195/195]
In [20]:
alignm=modeller.alignment(env)
In [21]:
alignm.append(file='P48816.fasta', align_codes='all',alignment_format='FASTA')
## создадим модель
mdl = modeller.model(env, file='1lmp.pdb', model_segment=('FIRST:'+'A', 'LAST:'+'A'))
## и добавим в выравнивание
alignm.append_model(mdl, atom_files='1lmp.pdb', align_codes='1lmp')
## есть смысл поправить идентификаторы
alignm[0].code = 'P48816'
In [22]:
alignm.salign()
alignm.write(file='all_in_one.ali', alignment_format='PIR')
SALIGN_____> adding the next group to the alignment; iteration 1
In [23]:
## Выбираем объект для моделирования
s = alignm[0]
pdb = alignm[1]
print s.code, pdb.code
## Создаем объект automodel
a = modeller.automodel.automodel(env, alnfile='all_in_one.ali', knowns= pdb.code, sequence = s.code )
a.name='mod'+s.code
a.starting_model = 1
a.ending_model = 2
a.make()
P48816 1lmp
automodel__W> Topology and/or parameter libraries already in memory. These will
be used instead of the automodel defaults. If this is not what you
want, clear them before creating the automodel object with
env.libs.topology.clear() and env.libs.parameters.clear()
fndatmi_285W> Only 129 residues out of 132 contain atoms of type CA
(This is usually caused by non-standard residues, such
as ligands, or by PDB files with missing atoms.)
fndatmi_285W> Only 129 residues out of 132 contain atoms of type CA
(This is usually caused by non-standard residues, such
as ligands, or by PDB files with missing atoms.)
check_ali___> Checking the sequence-structure alignment.
Implied target CA(i)-CA(i+1) distances longer than 8.0 angstroms:
ALN_POS TMPL RID1 RID2 NAM1 NAM2 DIST
----------------------------------------------
88 1 73 76 D C 8.895
END OF TABLE
getf_______W> RTF restraint not found in the atoms list:
residue type, indices: 2 137
atom names : C +N
atom indices : 1094 0
getf_______W> RTF restraint not found in the atoms list:
residue type, indices: 2 137
atom names : C CA +N O
atom indices : 1094 1091 0 1095
fndatmi_285W> Only 129 residues out of 132 contain atoms of type CA
(This is usually caused by non-standard residues, such
as ligands, or by PDB files with missing atoms.)
patch_s_522_> Number of disulfides patched in MODEL: 3
mdtrsr__446W> A potential that relies on one protein is used, yet you have at
least one known structure available. MDT, not library, potential is used.
iup2crm_280W> No topology library in memory or assigning a BLK residue.
Default CHARMM atom type assigned: C1 --> CT2
This message is written only for the first such atom.
0 atoms in HETATM residues constrained
to protein atoms within 2.30 angstroms
and protein CA atoms within 10.00 angstroms
0 atoms in residues without defined topology
constrained to be rigid bodies
condens_443_> Restraints marked for deletion were removed.
Total number of restraints before, now: 11705 10745
>> ENERGY; Differences between the model's features and restraints:
Number of all residues in MODEL : 137
Number of all, selected real atoms : 1096 1096
Number of all, selected pseudo atoms : 0 0
Number of all static, selected restraints : 10745 10745
COVALENT_CYS : F
NONBONDED_SEL_ATOMS : 1
Number of non-bonded pairs (excluding 1-2,1-3,1-4): 2233
Dynamic pairs routine : 2, NATM x NATM cell sorting
Atomic shift for contacts update (UPDATE_DYNAMIC) : 0.390
LENNARD_JONES_SWITCH : 6.500 7.500
COULOMB_JONES_SWITCH : 6.500 7.500
RESIDUE_SPAN_RANGE : 0 99999
NLOGN_USE : 15
CONTACT_SHELL : 4.000
DYNAMIC_PAIRS,_SPHERE,_COULOMB,_LENNARD,_MODELLER : T T F F F
SPHERE_STDV : 0.050
RADII_FACTOR : 0.820
Current energy : 1007.3779
Summary of the restraint violations:
NUM ... number of restraints.
NUMVI ... number of restraints with RVIOL > VIOL_REPORT_CUT[i].
RVIOL ... relative difference from the best value.
NUMVP ... number of restraints with -Ln(pdf) > VIOL_REPORT_CUT2[i].
RMS_1 ... RMS(feature, minimally_violated_basis_restraint, NUMB).
RMS_2 ... RMS(feature, best_value, NUMB).
MOL.PDF ... scaled contribution to -Ln(Molecular pdf).
# RESTRAINT_GROUP NUM NUMVI NUMVP RMS_1 RMS_2 MOL.PDF S_i
------------------------------------------------------------------------------------------------------
1 Bond length potential : 1119 0 2 0.009 0.009 25.128 1.000
2 Bond angle potential : 1504 1 9 2.192 2.192 145.96 1.000
3 Stereochemical cosine torsion poten: 713 0 31 47.795 47.795 259.86 1.000
4 Stereochemical improper torsion pot: 461 0 0 1.316 1.316 17.022 1.000
5 Soft-sphere overlap restraints : 2233 0 0 0.002 0.002 1.6206 1.000
6 Lennard-Jones 6-12 potential : 0 0 0 0.000 0.000 0.0000 1.000
7 Coulomb point-point electrostatic p: 0 0 0 0.000 0.000 0.0000 1.000
8 H-bonding potential : 0 0 0 0.000 0.000 0.0000 1.000
9 Distance restraints 1 (CA-CA) : 2215 0 4 0.305 0.305 100.59 1.000
10 Distance restraints 2 (N-O) : 2371 0 13 0.376 0.376 179.84 1.000
11 Mainchain Phi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
12 Mainchain Psi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
13 Mainchain Omega dihedral restraints: 136 0 3 4.655 4.655 34.750 1.000
14 Sidechain Chi_1 dihedral restraints: 121 0 2 72.319 72.319 31.006 1.000
15 Sidechain Chi_2 dihedral restraints: 87 0 0 78.507 78.507 40.041 1.000
16 Sidechain Chi_3 dihedral restraints: 35 0 0 79.937 79.937 21.786 1.000
17 Sidechain Chi_4 dihedral restraints: 22 0 0 85.796 85.796 16.488 1.000
18 Disulfide distance restraints : 3 0 0 0.029 0.029 0.44140 1.000
19 Disulfide angle restraints : 6 0 0 3.873 3.873 1.9871 1.000
20 Disulfide dihedral angle restraints: 3 0 0 33.456 33.456 2.7433 1.000
21 Lower bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
22 Upper bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
23 Distance restraints 3 (SDCH-MNCH) : 1316 0 0 0.355 0.355 28.599 1.000
24 Sidechain Chi_5 dihedral restraints: 0 0 0 0.000 0.000 0.0000 1.000
25 Phi/Psi pair of dihedral restraints: 135 19 15 30.106 63.202 76.594 1.000
26 Distance restraints 4 (SDCH-SDCH) : 498 0 0 0.537 0.537 22.931 1.000
27 Distance restraints 5 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
28 NMR distance restraints 6 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
29 NMR distance restraints 7 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
30 Minimal distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
31 Non-bonded restraints : 0 0 0 0.000 0.000 0.0000 1.000
32 Atomic accessibility restraints : 0 0 0 0.000 0.000 0.0000 1.000
33 Atomic density restraints : 0 0 0 0.000 0.000 0.0000 1.000
34 Absolute position restraints : 0 0 0 0.000 0.000 0.0000 1.000
35 Dihedral angle difference restraint: 0 0 0 0.000 0.000 0.0000 1.000
36 GBSA implicit solvent potential : 0 0 0 0.000 0.000 0.0000 1.000
37 EM density fitting potential : 0 0 0 0.000 0.000 0.0000 1.000
38 SAXS restraints : 0 0 0 0.000 0.000 0.0000 1.000
39 Symmetry restraints : 0 0 0 0.000 0.000 0.0000 1.000
# Heavy relative violation of each residue is written to: P48816.V99990001
# The profile is NOT normalized by the number of restraints.
# The profiles are smoothed over a window of residues: 1
# The sum of all numbers in the file: 15167.4219
List of the violated restraints:
A restraint is violated when the relative difference
from the best value (RVIOL) is larger than CUTOFF.
ICSR ... index of a restraint in the current set.
RESNO ... residue numbers of the first two atoms.
ATM ... IUPAC atom names of the first two atoms.
FEAT ... the value of the feature in the model.
restr ... the mean of the basis restraint with the smallest
difference from the model (local minimum).
viol ... difference from the local minimum.
rviol ... relative difference from the local minimum.
RESTR ... the best value (global minimum).
VIOL ... difference from the best value.
RVIOL ... relative difference from the best value.
-------------------------------------------------------------------------------------------------
Feature 2 : Bond angle potential
List of the RVIOL violations larger than : 4.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 2080 88D 88D N CA 697 698 124.76 107.00 17.76 5.11 107.00 17.76 5.11
-------------------------------------------------------------------------------------------------
Feature 25 : Phi/Psi pair of dihedral restraints
List of the RVIOL violations larger than : 6.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 3811 2Q 3K C N 16 18 -65.47 -70.20 13.44 0.86 -62.90 168.63 21.76
1 3K 3K N CA 18 19 127.81 140.40 -40.80
2 3812 3K 4L C N 25 27 -112.87 -108.50 23.08 1.29 -63.50 158.90 19.85
2 4L 4L N CA 27 28 109.84 132.50 -41.20
3 3815 6I 7F C N 49 51 -114.36 -124.20 21.17 1.16 -63.20 161.94 25.88
3 7F 7F N CA 51 52 162.05 143.30 -44.30
4 3816 7F 8A C N 60 62 -62.47 -68.20 29.14 2.15 -62.50 157.63 25.83
4 8A 8A N CA 62 63 116.73 145.30 -40.90
5 3817 8A 9L C N 65 67 -156.49 -108.50 49.40 2.23 -63.50 -162.23 33.20
5 9L 9L N CA 67 68 144.26 132.50 -41.20
6 3820 11V 12L C N 87 89 -61.61 -70.70 14.26 0.89 -63.50 171.83 23.97
6 12L 12L N CA 89 90 130.62 141.60 -41.20
7 3823 14V 15G C N 108 110 -69.72 -80.20 11.16 0.82 82.20 -132.45 6.74
7 15G 15G N CA 110 111 177.91 174.10 8.50
8 3824 15G 16S C N 112 114 -61.71 -72.40 34.68 1.71 -64.10 154.43 11.33
8 16S 16S N CA 114 115 119.41 152.40 -35.00
9 3844 35F 36E C N 274 276 58.74 54.60 12.02 0.65 -63.60 141.65 24.37
9 36E 36E N CA 276 277 31.11 42.40 -40.30
10 3860 51S 52S C N 411 413 64.49 56.90 9.69 0.67 -64.10 144.26 19.15
10 52S 52S N CA 413 414 30.38 36.40 -35.00
11 3893 84S 85P C N 675 677 -52.01 -58.70 54.54 3.99 -64.50 128.78 10.28
11 85P 85P N CA 677 678 -84.62 -30.50 147.20
12 3894 85P 86G C N 682 684 -66.25 -62.40 14.76 2.41 82.20 161.65 12.47
12 86G 86G N CA 684 685 -55.45 -41.20 8.50
13 3895 86G 87K C N 686 688 67.47 -70.20 137.84 10.22 -70.20 137.84 10.22
13 87K 87K N CA 688 689 147.28 140.40 140.40
14 3896 87K 88D C N 695 697 -153.84 -63.30 96.59 16.40 -63.30 96.59 16.40
14 88D 88D N CA 697 698 -73.63 -40.00 -40.00
15 3920 111I 112Y C N 872 874 -44.27 -98.40 55.36 2.55 -63.50 161.35 26.59
15 112Y 112Y N CA 874 875 116.80 128.40 -43.40
16 3934 125N 126H C N 1010 1012 -120.07 -63.20 69.12 8.42 -63.20 69.12 8.42
16 126H 126H N CA 1012 1013 -3.01 -42.30 -42.30
17 3935 126H 127C C N 1020 1022 -131.99 -63.00 72.94 10.64 -63.00 72.94 10.64
17 127C 127C N CA 1022 1023 -17.41 -41.10 -41.10
18 3936 127C 128Q C N 1026 1028 -59.93 -63.80 14.62 1.97 -121.10 176.81 9.14
18 128Q 128Q N CA 1028 1029 -54.41 -40.30 139.70
19 3937 128Q 129G C N 1035 1037 -73.74 -62.40 13.75 2.04 82.20 161.47 12.07
19 129G 129G N CA 1037 1038 -33.41 -41.20 8.50
report______> Distribution of short non-bonded contacts:
DISTANCE1: 0.00 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40
DISTANCE2: 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40 3.50
FREQUENCY: 0 0 0 0 0 9 10 54 77 107 113 114 144 171 181
<< end of ENERGY.
iupac_m_397W> Atoms were not swapped because of the uncertainty of how to handle the H atom.
>> ENERGY; Differences between the model's features and restraints:
Number of all residues in MODEL : 137
Number of all, selected real atoms : 1096 1096
Number of all, selected pseudo atoms : 0 0
Number of all static, selected restraints : 10745 10745
COVALENT_CYS : F
NONBONDED_SEL_ATOMS : 1
Number of non-bonded pairs (excluding 1-2,1-3,1-4): 2336
Dynamic pairs routine : 2, NATM x NATM cell sorting
Atomic shift for contacts update (UPDATE_DYNAMIC) : 0.390
LENNARD_JONES_SWITCH : 6.500 7.500
COULOMB_JONES_SWITCH : 6.500 7.500
RESIDUE_SPAN_RANGE : 0 99999
NLOGN_USE : 15
CONTACT_SHELL : 4.000
DYNAMIC_PAIRS,_SPHERE,_COULOMB,_LENNARD,_MODELLER : T T F F F
SPHERE_STDV : 0.050
RADII_FACTOR : 0.820
Current energy : 946.7557
Summary of the restraint violations:
NUM ... number of restraints.
NUMVI ... number of restraints with RVIOL > VIOL_REPORT_CUT[i].
RVIOL ... relative difference from the best value.
NUMVP ... number of restraints with -Ln(pdf) > VIOL_REPORT_CUT2[i].
RMS_1 ... RMS(feature, minimally_violated_basis_restraint, NUMB).
RMS_2 ... RMS(feature, best_value, NUMB).
MOL.PDF ... scaled contribution to -Ln(Molecular pdf).
# RESTRAINT_GROUP NUM NUMVI NUMVP RMS_1 RMS_2 MOL.PDF S_i
------------------------------------------------------------------------------------------------------
1 Bond length potential : 1119 0 2 0.009 0.009 24.320 1.000
2 Bond angle potential : 1504 1 7 2.127 2.127 138.40 1.000
3 Stereochemical cosine torsion poten: 713 0 29 47.960 47.960 257.85 1.000
4 Stereochemical improper torsion pot: 461 0 0 1.163 1.163 13.965 1.000
5 Soft-sphere overlap restraints : 2336 0 0 0.002 0.002 1.5842 1.000
6 Lennard-Jones 6-12 potential : 0 0 0 0.000 0.000 0.0000 1.000
7 Coulomb point-point electrostatic p: 0 0 0 0.000 0.000 0.0000 1.000
8 H-bonding potential : 0 0 0 0.000 0.000 0.0000 1.000
9 Distance restraints 1 (CA-CA) : 2215 0 2 0.310 0.310 97.928 1.000
10 Distance restraints 2 (N-O) : 2371 0 5 0.347 0.347 131.14 1.000
11 Mainchain Phi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
12 Mainchain Psi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
13 Mainchain Omega dihedral restraints: 136 0 2 4.337 4.337 30.167 1.000
14 Sidechain Chi_1 dihedral restraints: 121 0 1 74.298 74.298 33.677 1.000
15 Sidechain Chi_2 dihedral restraints: 87 0 0 69.940 69.940 33.402 1.000
16 Sidechain Chi_3 dihedral restraints: 35 0 0 71.655 71.655 24.796 1.000
17 Sidechain Chi_4 dihedral restraints: 22 0 0 111.147 111.147 17.999 1.000
18 Disulfide distance restraints : 3 0 0 0.024 0.024 0.30429 1.000
19 Disulfide angle restraints : 6 0 0 3.098 3.098 1.2717 1.000
20 Disulfide dihedral angle restraints: 3 0 0 33.141 33.141 2.6545 1.000
21 Lower bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
22 Upper bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
23 Distance restraints 3 (SDCH-MNCH) : 1316 0 0 0.331 0.331 25.246 1.000
24 Sidechain Chi_5 dihedral restraints: 0 0 0 0.000 0.000 0.0000 1.000
25 Phi/Psi pair of dihedral restraints: 135 16 19 30.646 56.773 83.880 1.000
26 Distance restraints 4 (SDCH-SDCH) : 498 0 0 0.597 0.597 28.170 1.000
27 Distance restraints 5 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
28 NMR distance restraints 6 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
29 NMR distance restraints 7 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
30 Minimal distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
31 Non-bonded restraints : 0 0 0 0.000 0.000 0.0000 1.000
32 Atomic accessibility restraints : 0 0 0 0.000 0.000 0.0000 1.000
33 Atomic density restraints : 0 0 0 0.000 0.000 0.0000 1.000
34 Absolute position restraints : 0 0 0 0.000 0.000 0.0000 1.000
35 Dihedral angle difference restraint: 0 0 0 0.000 0.000 0.0000 1.000
36 GBSA implicit solvent potential : 0 0 0 0.000 0.000 0.0000 1.000
37 EM density fitting potential : 0 0 0 0.000 0.000 0.0000 1.000
38 SAXS restraints : 0 0 0 0.000 0.000 0.0000 1.000
39 Symmetry restraints : 0 0 0 0.000 0.000 0.0000 1.000
# Heavy relative violation of each residue is written to: P48816.V99990002
# The profile is NOT normalized by the number of restraints.
# The profiles are smoothed over a window of residues: 1
# The sum of all numbers in the file: 14697.0605
List of the violated restraints:
A restraint is violated when the relative difference
from the best value (RVIOL) is larger than CUTOFF.
ICSR ... index of a restraint in the current set.
RESNO ... residue numbers of the first two atoms.
ATM ... IUPAC atom names of the first two atoms.
FEAT ... the value of the feature in the model.
restr ... the mean of the basis restraint with the smallest
difference from the model (local minimum).
viol ... difference from the local minimum.
rviol ... relative difference from the local minimum.
RESTR ... the best value (global minimum).
VIOL ... difference from the best value.
RVIOL ... relative difference from the best value.
-------------------------------------------------------------------------------------------------
Feature 2 : Bond angle potential
List of the RVIOL violations larger than : 4.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 2080 88D 88D N CA 697 698 127.94 107.00 20.94 6.02 107.00 20.94 6.02
-------------------------------------------------------------------------------------------------
Feature 25 : Phi/Psi pair of dihedral restraints
List of the RVIOL violations larger than : 6.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 3810 1M 2Q C N 7 9 -108.19 -121.10 38.69 2.00 -63.80 150.23 24.46
1 2Q 2Q N CA 9 10 176.17 139.70 -40.30
2 3812 3K 4L C N 25 27 -83.92 -70.70 14.25 1.09 -63.50 173.10 25.25
2 4L 4L N CA 27 28 146.91 141.60 -41.20
3 3815 6I 7F C N 49 51 -165.43 -124.20 52.29 1.36 -63.20 173.54 30.31
3 7F 7F N CA 51 52 175.47 143.30 -44.30
4 3816 7F 8A C N 60 62 -72.87 -68.20 14.80 1.03 -62.50 160.10 26.76
4 8A 8A N CA 62 63 159.34 145.30 -40.90
5 3817 8A 9L C N 65 67 -65.30 -70.70 7.94 0.79 -63.50 171.38 23.90
5 9L 9L N CA 67 68 147.43 141.60 -41.20
6 3821 12L 13C C N 95 97 -63.17 -63.00 3.38 0.42 -117.90 -177.19 7.87
6 13C 13C N CA 97 98 -44.47 -41.10 141.10
7 3823 14V 15G C N 108 110 76.98 78.70 4.99 0.21 82.20 179.37 8.97
7 15G 15G N CA 110 111 -170.79 -166.10 8.50
8 3824 15G 16S C N 112 114 -122.18 -136.60 34.42 1.38 -64.10 165.47 10.21
8 16S 16S N CA 114 115 119.94 151.20 -35.00
9 3844 35F 36E C N 274 276 54.64 54.60 6.82 0.52 -63.60 140.49 24.17
9 36E 36E N CA 276 277 35.58 42.40 -40.30
10 3859 50E 51S C N 405 407 -138.29 -64.10 84.82 8.56 -64.10 84.82 8.56
10 51S 51S N CA 407 408 6.12 -35.00 -35.00
11 3860 51S 52S C N 411 413 60.05 56.90 5.17 0.26 -64.10 141.22 18.68
11 52S 52S N CA 413 414 32.30 36.40 -35.00
12 3896 87K 88D C N 695 697 -143.48 -63.30 82.74 13.88 -63.30 82.74 13.88
12 88D 88D N CA 697 698 -60.44 -40.00 -40.00
13 3922 113K 114R C N 893 895 -52.71 -125.20 77.18 2.47 -63.00 155.54 21.71
13 114R 114R N CA 895 896 114.11 140.60 -41.10
14 3923 114R 115H C N 904 906 154.21 -63.20 142.66 22.42 -63.20 142.66 22.42
14 115H 115H N CA 906 907 -37.94 -42.30 -42.30
15 3934 125N 126H C N 1010 1012 -119.50 -63.20 72.63 8.52 -63.20 72.63 8.52
15 126H 126H N CA 1012 1013 3.58 -42.30 -42.30
16 3935 126H 127C C N 1020 1022 -119.86 -63.00 56.94 9.34 -63.00 56.94 9.34
16 127C 127C N CA 1022 1023 -38.18 -41.10 -41.10
report______> Distribution of short non-bonded contacts:
DISTANCE1: 0.00 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40
DISTANCE2: 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40 3.50
FREQUENCY: 0 0 0 0 0 15 17 66 76 110 113 132 149 188 172
<< end of ENERGY.
>> Summary of successfully produced models:
Filename molpdf
----------------------------------------
P48816.B99990001.pdb 1007.37793
P48816.B99990002.pdb 946.75574
In [24]:
import nglview
import ipywidgets
w1 = nglview.show_structure_file('P48816.B99990001.pdb')
w1
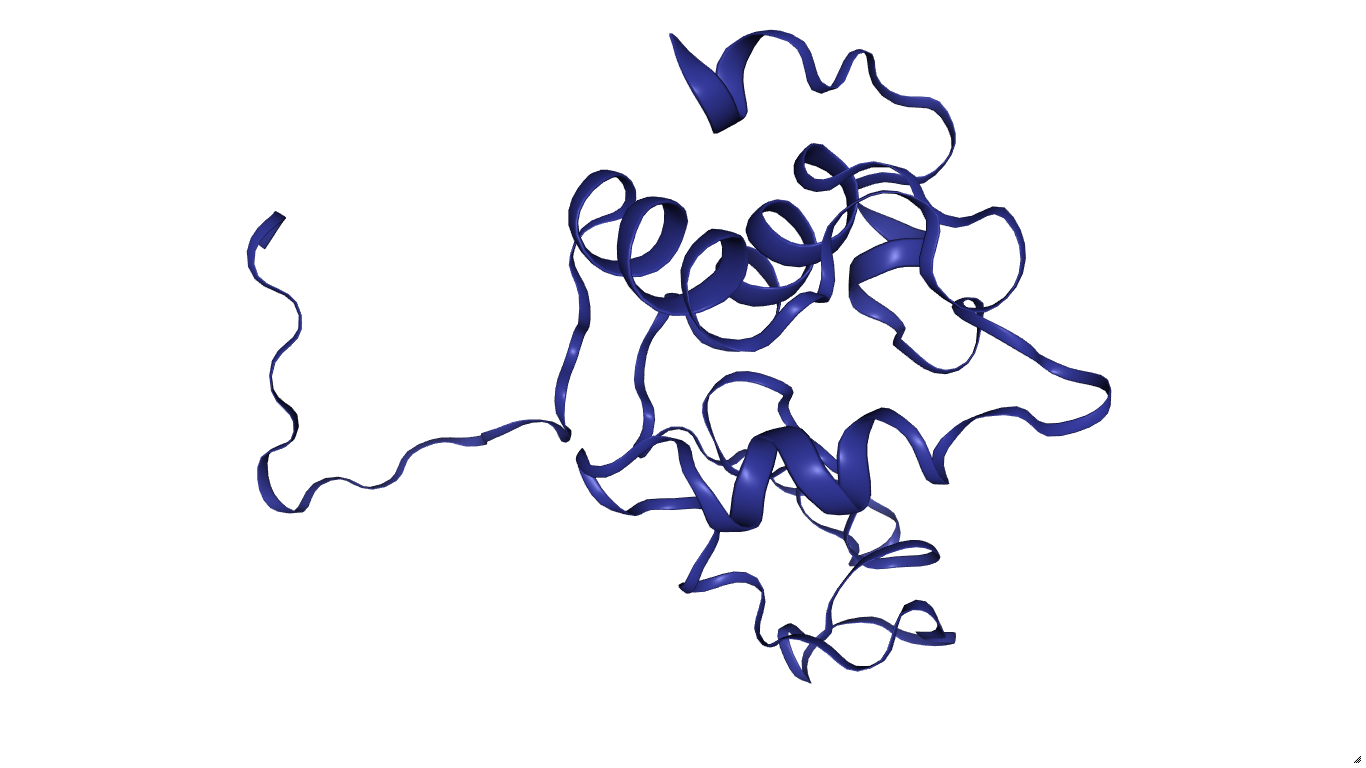
Стоит отметить, что белок тут без лиганда. Действительно, мы же его не добавили. Поэтому добавим три остатка лиганда в последовательности (смотри три последних остатка в pdb файле).
In [26]:
## Получим список остатков последовательности
str_res=""
for res in alignm[0].residues:
str_res=str_res+res.code #берем три последник остатка - лиганды
str_res=str_res+"..."
print str_res
alignm[0].code="bommo"
MQKLIIFALVVLCVGSEAKTFTRCGLVHELRKHGFEENLMRNWVCLVEHESSRDTSKTNTNRNGSKDYGLFQINDRYWCSKGASPGKDCNVKCSDLLTDDITKAAKCAKKIYKRHRFDAWYGWKNHCQGSLPDISSC...
In [30]:
## Добавим в объект выравнивание последовательность из строки
alignm.append_sequence(str_res)
alignm[2].code="with_lig"
In [31]:
alignm.salign()
alignm.write(file='all_in_one.ali', alignment_format='PIR')
SALIGN_____> adding the next group to the alignment; iteration 1 SALIGN_____> adding the next group to the alignment; iteration 2 SALIGN_____> adding the next group to the alignment; iteration 3
In [32]:
## Выбираем объект для моделирования
new = alignm[2]
pdb = alignm[1]
print pdb.code,new.code
## Создаем объект automodel
a = modeller.automodel.automodel(env, alnfile='all_in_one.ali', knowns= pdb.code, sequence = new.code )
a.name='mod'+s.code
a.starting_model = 1
a.ending_model = 2
a.make()
1lmp with_lig
automodel__W> Topology and/or parameter libraries already in memory. These will
be used instead of the automodel defaults. If this is not what you
want, clear them before creating the automodel object with
env.libs.topology.clear() and env.libs.parameters.clear()
fndatmi_285W> Only 129 residues out of 132 contain atoms of type CA
(This is usually caused by non-standard residues, such
as ligands, or by PDB files with missing atoms.)
fndatmi_285W> Only 129 residues out of 132 contain atoms of type CA
(This is usually caused by non-standard residues, such
as ligands, or by PDB files with missing atoms.)
check_ali___> Checking the sequence-structure alignment.
Implied target CA(i)-CA(i+1) distances longer than 8.0 angstroms:
ALN_POS TMPL RID1 RID2 NAM1 NAM2 DIST
----------------------------------------------
88 1 73 76 D C 8.895
END OF TABLE
getf_______W> RTF restraint not found in the atoms list:
residue type, indices: 2 137
atom names : C +N
atom indices : 1094 0
getf_______W> RTF restraint not found in the atoms list:
residue type, indices: 2 137
atom names : C CA +N O
atom indices : 1094 1091 0 1095
fndatmi_285W> Only 129 residues out of 132 contain atoms of type CA
(This is usually caused by non-standard residues, such
as ligands, or by PDB files with missing atoms.)
patch_s_522_> Number of disulfides patched in MODEL: 3
mdtrsr__446W> A potential that relies on one protein is used, yet you have at
least one known structure available. MDT, not library, potential is used.
iup2crm_280W> No topology library in memory or assigning a BLK residue.
Default CHARMM atom type assigned: C1 --> CT2
This message is written only for the first such atom.
43 atoms in HETATM residues constrained
to protein atoms within 2.30 angstroms
and protein CA atoms within 10.00 angstroms
43 atoms in residues without defined topology
constrained to be rigid bodies
condens_443_> Restraints marked for deletion were removed.
Total number of restraints before, now: 13094 12134
read_pd_403W> Treating residue type NAG as a BLK (rigid body)
even though topology information appears to be at least
partially available. To treat this residue flexibly
instead, remove the corresponding 'MODELLER BLK RESIDUE'
REMARK from the input PDB file.
read_pd_403W> Treating residue type NAG as a BLK (rigid body)
even though topology information appears to be at least
partially available. To treat this residue flexibly
instead, remove the corresponding 'MODELLER BLK RESIDUE'
REMARK from the input PDB file.
read_pd_403W> Treating residue type NDG as a BLK (rigid body)
even though topology information appears to be at least
partially available. To treat this residue flexibly
instead, remove the corresponding 'MODELLER BLK RESIDUE'
REMARK from the input PDB file.
>> ENERGY; Differences between the model's features and restraints:
Number of all residues in MODEL : 140
Number of all, selected real atoms : 1139 1139
Number of all, selected pseudo atoms : 0 0
Number of all static, selected restraints : 12134 12134
COVALENT_CYS : F
NONBONDED_SEL_ATOMS : 1
Number of non-bonded pairs (excluding 1-2,1-3,1-4): 2529
Dynamic pairs routine : 2, NATM x NATM cell sorting
Atomic shift for contacts update (UPDATE_DYNAMIC) : 0.390
LENNARD_JONES_SWITCH : 6.500 7.500
COULOMB_JONES_SWITCH : 6.500 7.500
RESIDUE_SPAN_RANGE : 0 99999
NLOGN_USE : 15
CONTACT_SHELL : 4.000
DYNAMIC_PAIRS,_SPHERE,_COULOMB,_LENNARD,_MODELLER : T T F F F
SPHERE_STDV : 0.050
RADII_FACTOR : 0.820
Current energy : 1064.6942
Summary of the restraint violations:
NUM ... number of restraints.
NUMVI ... number of restraints with RVIOL > VIOL_REPORT_CUT[i].
RVIOL ... relative difference from the best value.
NUMVP ... number of restraints with -Ln(pdf) > VIOL_REPORT_CUT2[i].
RMS_1 ... RMS(feature, minimally_violated_basis_restraint, NUMB).
RMS_2 ... RMS(feature, best_value, NUMB).
MOL.PDF ... scaled contribution to -Ln(Molecular pdf).
# RESTRAINT_GROUP NUM NUMVI NUMVP RMS_1 RMS_2 MOL.PDF S_i
------------------------------------------------------------------------------------------------------
1 Bond length potential : 1119 0 5 0.009 0.009 28.462 1.000
2 Bond angle potential : 1504 1 14 2.338 2.338 164.61 1.000
3 Stereochemical cosine torsion poten: 713 0 28 48.242 48.242 259.30 1.000
4 Stereochemical improper torsion pot: 461 0 0 1.310 1.310 17.624 1.000
5 Soft-sphere overlap restraints : 2529 1 2 0.007 0.007 16.480 1.000
6 Lennard-Jones 6-12 potential : 0 0 0 0.000 0.000 0.0000 1.000
7 Coulomb point-point electrostatic p: 0 0 0 0.000 0.000 0.0000 1.000
8 H-bonding potential : 0 0 0 0.000 0.000 0.0000 1.000
9 Distance restraints 1 (CA-CA) : 2215 0 3 0.303 0.303 99.203 1.000
10 Distance restraints 2 (N-O) : 2371 0 11 0.344 0.344 153.50 1.000
11 Mainchain Phi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
12 Mainchain Psi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
13 Mainchain Omega dihedral restraints: 136 0 4 5.024 5.024 40.481 1.000
14 Sidechain Chi_1 dihedral restraints: 121 0 2 81.442 81.442 39.708 1.000
15 Sidechain Chi_2 dihedral restraints: 87 0 0 73.920 73.920 38.910 1.000
16 Sidechain Chi_3 dihedral restraints: 35 0 0 83.022 83.022 24.050 1.000
17 Sidechain Chi_4 dihedral restraints: 22 0 0 99.021 99.021 14.391 1.000
18 Disulfide distance restraints : 3 0 0 0.023 0.023 0.27864 1.000
19 Disulfide angle restraints : 6 0 0 2.646 2.646 0.92721 1.000
20 Disulfide dihedral angle restraints: 3 0 0 22.772 22.772 1.5937 1.000
21 Lower bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
22 Upper bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
23 Distance restraints 3 (SDCH-MNCH) : 1316 0 0 0.344 0.344 24.366 1.000
24 Sidechain Chi_5 dihedral restraints: 0 0 0 0.000 0.000 0.0000 1.000
25 Phi/Psi pair of dihedral restraints: 135 17 17 29.043 65.341 87.747 1.000
26 Distance restraints 4 (SDCH-SDCH) : 498 0 0 0.566 0.566 25.649 1.000
27 Distance restraints 5 (X-Y) : 1389 0 1 0.050 0.050 27.419 1.000
28 NMR distance restraints 6 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
29 NMR distance restraints 7 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
30 Minimal distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
31 Non-bonded restraints : 0 0 0 0.000 0.000 0.0000 1.000
32 Atomic accessibility restraints : 0 0 0 0.000 0.000 0.0000 1.000
33 Atomic density restraints : 0 0 0 0.000 0.000 0.0000 1.000
34 Absolute position restraints : 0 0 0 0.000 0.000 0.0000 1.000
35 Dihedral angle difference restraint: 0 0 0 0.000 0.000 0.0000 1.000
36 GBSA implicit solvent potential : 0 0 0 0.000 0.000 0.0000 1.000
37 EM density fitting potential : 0 0 0 0.000 0.000 0.0000 1.000
38 SAXS restraints : 0 0 0 0.000 0.000 0.0000 1.000
39 Symmetry restraints : 0 0 0 0.000 0.000 0.0000 1.000
# Heavy relative violation of each residue is written to: with_lig.V99990001
# The profile is NOT normalized by the number of restraints.
# The profiles are smoothed over a window of residues: 1
# The sum of all numbers in the file: 16211.8447
List of the violated restraints:
A restraint is violated when the relative difference
from the best value (RVIOL) is larger than CUTOFF.
ICSR ... index of a restraint in the current set.
RESNO ... residue numbers of the first two atoms.
ATM ... IUPAC atom names of the first two atoms.
FEAT ... the value of the feature in the model.
restr ... the mean of the basis restraint with the smallest
difference from the model (local minimum).
viol ... difference from the local minimum.
rviol ... relative difference from the local minimum.
RESTR ... the best value (global minimum).
VIOL ... difference from the best value.
RVIOL ... relative difference from the best value.
-------------------------------------------------------------------------------------------------
Feature 2 : Bond angle potential
List of the RVIOL violations larger than : 4.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 2080 88D 88D N CA 697 698 129.61 107.00 22.61 6.50 107.00 22.61 6.50
-------------------------------------------------------------------------------------------------
Feature 25 : Phi/Psi pair of dihedral restraints
List of the RVIOL violations larger than : 6.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 3810 1M 2Q C N 7 9 -77.54 -73.00 21.56 1.32 -63.80 158.52 24.04
1 2Q 2Q N CA 9 10 161.78 140.70 -40.30
2 3811 2Q 3K C N 16 18 -62.20 -70.20 12.88 0.77 -62.90 171.11 22.26
2 3K 3K N CA 18 19 130.31 140.40 -40.80
3 3812 3K 4L C N 25 27 -93.25 -108.50 18.50 1.01 -63.50 178.32 26.56
3 4L 4L N CA 27 28 142.97 132.50 -41.20
4 3815 6I 7F C N 49 51 -95.67 -71.40 24.85 1.85 -63.20 -177.39 23.26
4 7F 7F N CA 51 52 135.40 140.70 -44.30
5 3816 7F 8A C N 60 62 -125.55 -134.00 10.21 0.25 -62.50 -171.31 34.29
5 8A 8A N CA 62 63 141.26 147.00 -40.90
6 3817 8A 9L C N 65 67 -113.04 -108.50 8.76 0.42 -63.50 -174.46 28.81
6 9L 9L N CA 67 68 139.99 132.50 -41.20
7 3820 11V 12L C N 87 89 -64.58 -70.70 9.22 0.58 -63.50 175.91 24.36
7 12L 12L N CA 89 90 134.71 141.60 -41.20
8 3823 14V 15G C N 108 110 -84.84 -80.20 4.91 0.26 82.20 -123.65 7.39
8 15G 15G N CA 110 111 175.72 174.10 8.50
9 3824 15G 16S C N 112 114 -67.28 -72.40 21.87 1.12 -64.10 166.17 11.85
9 16S 16S N CA 114 115 131.14 152.40 -35.00
10 3844 35F 36E C N 274 276 52.88 54.60 5.30 0.54 -63.60 140.01 24.08
10 36E 36E N CA 276 277 37.38 42.40 -40.30
11 3860 51S 52S C N 411 413 63.53 56.90 8.91 0.57 -64.10 143.44 19.03
11 52S 52S N CA 413 414 30.45 36.40 -35.00
12 3896 87K 88D C N 695 697 169.10 -96.50 96.59 3.93 -63.30 -175.13 20.70
12 88D 88D N CA 697 698 93.78 114.20 -40.00
13 3922 113K 114R C N 893 895 -63.09 -125.20 76.79 2.27 -63.00 136.54 18.44
13 114R 114R N CA 895 896 95.44 140.60 -41.10
14 3923 114R 115H C N 904 906 178.11 -63.20 118.71 18.74 -63.20 118.71 18.74
14 115H 115H N CA 906 907 -40.30 -42.30 -42.30
15 3935 126H 127C C N 1020 1022 -122.02 -63.00 63.46 9.04 -63.00 63.46 9.04
15 127C 127C N CA 1022 1023 -17.75 -41.10 -41.10
16 3936 127C 128Q C N 1026 1028 -60.03 -63.80 13.21 1.77 -121.10 178.14 9.21
16 128Q 128Q N CA 1028 1029 -52.96 -40.30 139.70
17 3937 128Q 129G C N 1035 1037 -73.89 -62.40 13.47 2.04 82.20 161.82 12.11
17 129G 129G N CA 1037 1038 -34.18 -41.20 8.50
report______> Distribution of short non-bonded contacts:
DISTANCE1: 0.00 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40
DISTANCE2: 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40 3.50
FREQUENCY: 0 0 0 0 0 11 16 65 92 129 130 142 161 208 177
<< end of ENERGY.
read_pd_403W> Treating residue type NAG as a BLK (rigid body)
even though topology information appears to be at least
partially available. To treat this residue flexibly
instead, remove the corresponding 'MODELLER BLK RESIDUE'
REMARK from the input PDB file.
read_pd_403W> Treating residue type NAG as a BLK (rigid body)
even though topology information appears to be at least
partially available. To treat this residue flexibly
instead, remove the corresponding 'MODELLER BLK RESIDUE'
REMARK from the input PDB file.
read_pd_403W> Treating residue type NDG as a BLK (rigid body)
even though topology information appears to be at least
partially available. To treat this residue flexibly
instead, remove the corresponding 'MODELLER BLK RESIDUE'
REMARK from the input PDB file.
iupac_m_397W> Atoms were not swapped because of the uncertainty of how to handle the H atom.
iupac_m_397W> Atoms were not swapped because of the uncertainty of how to handle the H atom.
iupac_m_397W> Atoms were not swapped because of the uncertainty of how to handle the H atom.
>> ENERGY; Differences between the model's features and restraints:
Number of all residues in MODEL : 140
Number of all, selected real atoms : 1139 1139
Number of all, selected pseudo atoms : 0 0
Number of all static, selected restraints : 12134 12134
COVALENT_CYS : F
NONBONDED_SEL_ATOMS : 1
Number of non-bonded pairs (excluding 1-2,1-3,1-4): 2413
Dynamic pairs routine : 2, NATM x NATM cell sorting
Atomic shift for contacts update (UPDATE_DYNAMIC) : 0.390
LENNARD_JONES_SWITCH : 6.500 7.500
COULOMB_JONES_SWITCH : 6.500 7.500
RESIDUE_SPAN_RANGE : 0 99999
NLOGN_USE : 15
CONTACT_SHELL : 4.000
DYNAMIC_PAIRS,_SPHERE,_COULOMB,_LENNARD,_MODELLER : T T F F F
SPHERE_STDV : 0.050
RADII_FACTOR : 0.820
Current energy : 1164.5482
Summary of the restraint violations:
NUM ... number of restraints.
NUMVI ... number of restraints with RVIOL > VIOL_REPORT_CUT[i].
RVIOL ... relative difference from the best value.
NUMVP ... number of restraints with -Ln(pdf) > VIOL_REPORT_CUT2[i].
RMS_1 ... RMS(feature, minimally_violated_basis_restraint, NUMB).
RMS_2 ... RMS(feature, best_value, NUMB).
MOL.PDF ... scaled contribution to -Ln(Molecular pdf).
# RESTRAINT_GROUP NUM NUMVI NUMVP RMS_1 RMS_2 MOL.PDF S_i
------------------------------------------------------------------------------------------------------
1 Bond length potential : 1119 0 2 0.010 0.010 31.949 1.000
2 Bond angle potential : 1504 1 12 2.401 2.401 174.25 1.000
3 Stereochemical cosine torsion poten: 713 0 29 47.625 47.625 253.54 1.000
4 Stereochemical improper torsion pot: 461 0 0 1.528 1.528 22.864 1.000
5 Soft-sphere overlap restraints : 2413 1 2 0.008 0.008 16.405 1.000
6 Lennard-Jones 6-12 potential : 0 0 0 0.000 0.000 0.0000 1.000
7 Coulomb point-point electrostatic p: 0 0 0 0.000 0.000 0.0000 1.000
8 H-bonding potential : 0 0 0 0.000 0.000 0.0000 1.000
9 Distance restraints 1 (CA-CA) : 2215 0 4 0.295 0.295 103.58 1.000
10 Distance restraints 2 (N-O) : 2371 3 12 0.365 0.365 181.62 1.000
11 Mainchain Phi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
12 Mainchain Psi dihedral restraints : 0 0 0 0.000 0.000 0.0000 1.000
13 Mainchain Omega dihedral restraints: 136 0 2 4.716 4.716 35.666 1.000
14 Sidechain Chi_1 dihedral restraints: 121 0 1 79.453 79.453 53.180 1.000
15 Sidechain Chi_2 dihedral restraints: 87 0 1 75.268 75.268 36.806 1.000
16 Sidechain Chi_3 dihedral restraints: 35 0 0 83.944 83.944 23.705 1.000
17 Sidechain Chi_4 dihedral restraints: 22 0 0 89.489 89.489 15.271 1.000
18 Disulfide distance restraints : 3 0 0 0.029 0.029 0.44171 1.000
19 Disulfide angle restraints : 6 0 0 3.039 3.039 1.2239 1.000
20 Disulfide dihedral angle restraints: 3 0 0 22.033 22.033 1.5137 1.000
21 Lower bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
22 Upper bound distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
23 Distance restraints 3 (SDCH-MNCH) : 1316 0 0 0.391 0.391 37.258 1.000
24 Sidechain Chi_5 dihedral restraints: 0 0 0 0.000 0.000 0.0000 1.000
25 Phi/Psi pair of dihedral restraints: 135 17 19 28.525 58.349 101.70 1.000
26 Distance restraints 4 (SDCH-SDCH) : 498 0 2 0.769 0.769 49.054 1.000
27 Distance restraints 5 (X-Y) : 1389 0 0 0.046 0.046 24.525 1.000
28 NMR distance restraints 6 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
29 NMR distance restraints 7 (X-Y) : 0 0 0 0.000 0.000 0.0000 1.000
30 Minimal distance restraints : 0 0 0 0.000 0.000 0.0000 1.000
31 Non-bonded restraints : 0 0 0 0.000 0.000 0.0000 1.000
32 Atomic accessibility restraints : 0 0 0 0.000 0.000 0.0000 1.000
33 Atomic density restraints : 0 0 0 0.000 0.000 0.0000 1.000
34 Absolute position restraints : 0 0 0 0.000 0.000 0.0000 1.000
35 Dihedral angle difference restraint: 0 0 0 0.000 0.000 0.0000 1.000
36 GBSA implicit solvent potential : 0 0 0 0.000 0.000 0.0000 1.000
37 EM density fitting potential : 0 0 0 0.000 0.000 0.0000 1.000
38 SAXS restraints : 0 0 0 0.000 0.000 0.0000 1.000
39 Symmetry restraints : 0 0 0 0.000 0.000 0.0000 1.000
# Heavy relative violation of each residue is written to: with_lig.V99990002
# The profile is NOT normalized by the number of restraints.
# The profiles are smoothed over a window of residues: 1
# The sum of all numbers in the file: 16359.2695
List of the violated restraints:
A restraint is violated when the relative difference
from the best value (RVIOL) is larger than CUTOFF.
ICSR ... index of a restraint in the current set.
RESNO ... residue numbers of the first two atoms.
ATM ... IUPAC atom names of the first two atoms.
FEAT ... the value of the feature in the model.
restr ... the mean of the basis restraint with the smallest
difference from the model (local minimum).
viol ... difference from the local minimum.
rviol ... relative difference from the local minimum.
RESTR ... the best value (global minimum).
VIOL ... difference from the best value.
RVIOL ... relative difference from the best value.
-------------------------------------------------------------------------------------------------
Feature 2 : Bond angle potential
List of the RVIOL violations larger than : 4.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 2080 88D 88D N CA 697 698 128.58 107.00 21.58 6.21 107.00 21.58 6.21
-------------------------------------------------------------------------------------------------
Feature 10 : Distance restraints 2 (N-O)
List of the RVIOL violations larger than : 4.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 8023 86G 63N N O 684 508 10.74 7.27 3.46 4.89 7.27 3.46 4.89
2 8033 87K 63N N O 688 508 12.09 8.66 3.43 5.73 8.66 3.43 5.73
-------------------------------------------------------------------------------------------------
Feature 25 : Phi/Psi pair of dihedral restraints
List of the RVIOL violations larger than : 6.5000
# ICSR RESNO1/2 ATM1/2 INDATM1/2 FEAT restr viol rviol RESTR VIOL RVIOL
1 3810 1M 2Q C N 7 9 -135.73 -121.10 69.00 3.01 -63.80 133.59 23.29
1 2Q 2Q N CA 9 10 -152.87 139.70 -40.30
2 3811 2Q 3K C N 16 18 -57.28 -70.20 20.60 1.24 -62.90 165.26 21.77
2 3K 3K N CA 18 19 124.36 140.40 -40.80
3 3812 3K 4L C N 25 27 -81.13 -70.70 10.45 0.93 -63.50 177.55 25.70
3 4L 4L N CA 27 28 142.13 141.60 -41.20
4 3815 6I 7F C N 49 51 -144.90 -124.20 64.45 2.46 -63.20 138.11 24.13
4 7F 7F N CA 51 52 -155.66 143.30 -44.30
5 3816 7F 8A C N 60 62 -62.40 -68.20 28.79 2.52 -62.50 145.60 23.86
5 8A 8A N CA 62 63 173.50 145.30 -40.90
6 3820 11V 12L C N 87 89 -101.79 -108.50 41.38 2.16 -63.50 138.28 17.41
6 12L 12L N CA 89 90 91.67 132.50 -41.20
7 3821 12L 13C C N 95 97 -66.40 -63.00 4.02 0.52 -117.90 -172.83 8.02
7 13C 13C N CA 97 98 -38.95 -41.10 141.10
8 3823 14V 15G C N 108 110 -99.30 -80.20 34.54 0.85 82.20 -116.50 7.95
8 15G 15G N CA 110 111 -157.12 174.10 8.50
9 3824 15G 16S C N 112 114 -58.95 -72.40 37.38 1.80 -64.10 152.61 11.37
9 16S 16S N CA 114 115 117.52 152.40 -35.00
10 3844 35F 36E C N 274 276 58.76 54.60 14.38 0.81 -63.60 140.45 24.16
10 36E 36E N CA 276 277 28.64 42.40 -40.30
11 3859 50E 51S C N 405 407 -136.03 -64.10 83.44 8.27 -64.10 83.44 8.27
11 51S 51S N CA 407 408 7.29 -35.00 -35.00
12 3860 51S 52S C N 411 413 61.42 56.90 6.66 0.38 -64.10 142.05 18.81
12 52S 52S N CA 413 414 31.50 36.40 -35.00
13 3871 62R 63N C N 499 501 -77.43 -119.90 64.83 3.52 -63.20 133.68 17.51
13 63N 63N N CA 501 502 -174.02 137.00 -41.10
14 3872 63N 64G C N 507 509 -65.62 -62.40 24.74 3.55 82.20 149.95 10.87
14 64G 64G N CA 509 510 -16.67 -41.20 8.50
15 3896 87K 88D C N 695 697 -145.75 -63.30 85.84 14.46 -63.30 85.84 14.46
15 88D 88D N CA 697 698 -63.86 -40.00 -40.00
16 3919 110K 111I C N 864 866 -57.40 -63.40 42.73 7.22 -63.40 42.73 7.22
16 111I 111I N CA 866 867 -1.30 -43.60 -43.60
17 3935 126H 127C C N 1020 1022 -117.74 -63.00 56.25 8.59 -63.00 56.25 8.59
17 127C 127C N CA 1022 1023 -28.14 -41.10 -41.10
report______> Distribution of short non-bonded contacts:
DISTANCE1: 0.00 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40
DISTANCE2: 2.10 2.20 2.30 2.40 2.50 2.60 2.70 2.80 2.90 3.00 3.10 3.20 3.30 3.40 3.50
FREQUENCY: 0 0 0 0 1 9 12 65 70 125 127 143 167 198 183
<< end of ENERGY.
>> Summary of successfully produced models:
Filename molpdf
----------------------------------------
with_lig.B99990001.pdb 1064.69421
with_lig.B99990002.pdb 1164.54822
In [34]:
import nglview
import ipywidgets
w1 = nglview.show_structure_file('with_lig.B99990001.pdb')
w1
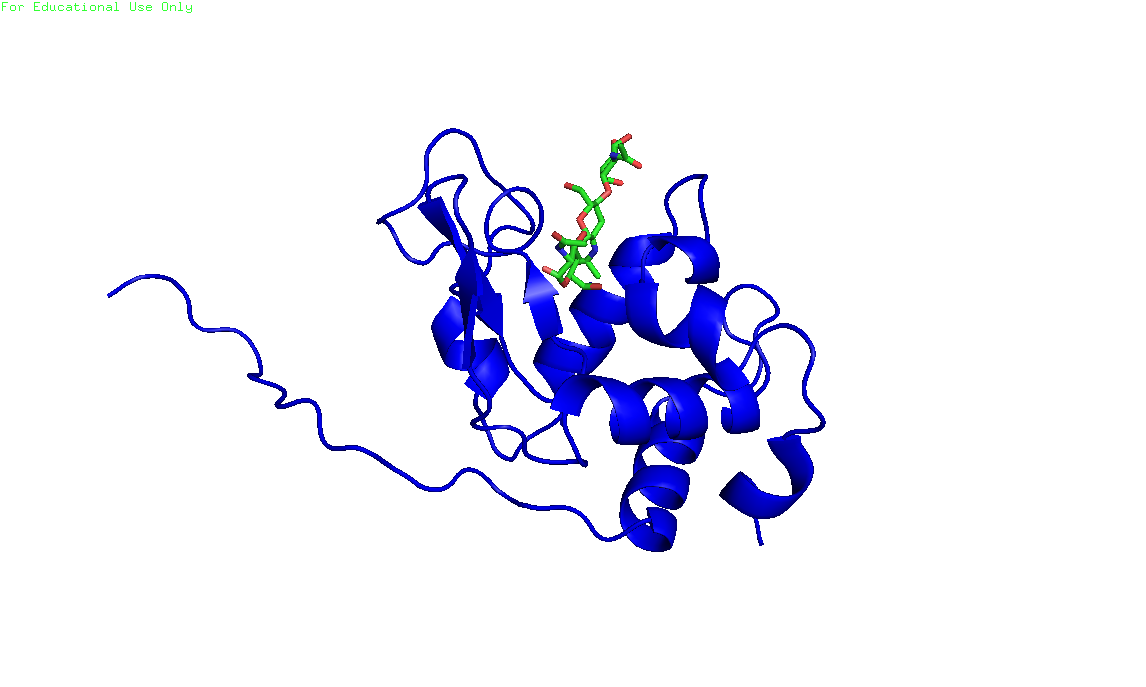
In [ ]: