Figure 1. Multiple alignment
Download link
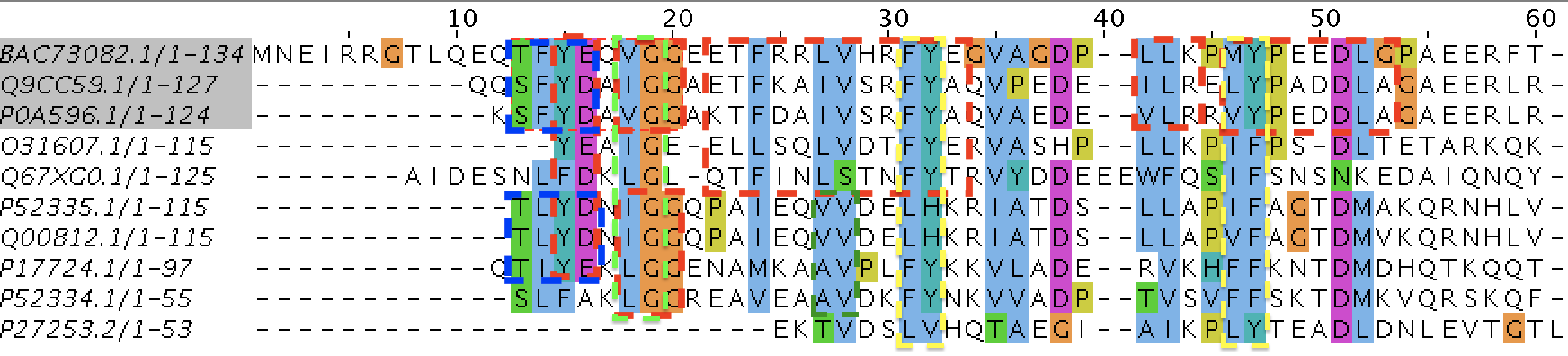
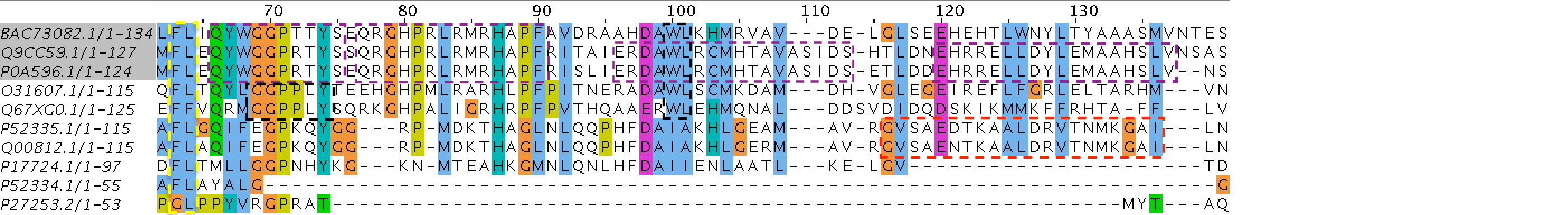
Used programs: BLAST
Set parameters:
Table 1. Hit table results
# Name Subject ids % Identity % Positives AlignLen Mismatches Gap opens Q. start Q. end S. start S. end E-value Bit score % Coverage Homology 1
Group 2 truncated hemoglobin GlbO
gi|17366693|sp|Q9CC59.1|TRHBO_MYCLE
53.543
66.14
127
56
1
11
134
2
128
1.77e-42
139
99.2
+
2
Group 2 truncated hemoglobin GlbO
gi|61224558|sp|P0A596.1|TRHBO_MYCBO;gi|615880539|sp|P9WN22.1|TRHBO_MYCTO;gi|615880555|sp|P9WN23.1|TRHBO_MYCTU
55.645
68.55
124
52
1
12
132
3
126
2.36e-42
139
96.9
+
3
Group 2 truncated hemoglobin
gi|81341886|sp|O31607.1|TRHBO_BACSU
46.154
56.41
117
61
2
15
131
9
123
9.09e-26
97.1
88.6
+
4
Two-on-two hemoglobin-3
gi|75322687|sp|Q67XG0.1|GLB3_ARATH
27.778
50.79
126
87
3
8
130
21
145
1.92e-10
58.5
72
+
5
Group 1 truncated hemoglobin
gi|1707915|sp|P52335.1|TRHBN_NOSSN
26.050
45.38
119
84
2
13
131
3
117
9.75e-09
52.8
100.8
+
6
Group 1 truncated hemoglobin
gi|232163|sp|Q00812.1|TRHBN_NOSCO
27.273
47.11
121
80
4
13
131
3
117
4.66e-08
51.2
102.5
+
7
Group 1 truncated hemoglobin
gi|121274|sp|P17724.1|TRHBN_TETPY
26.923
48.08
104
66
2
12
112
5
101
4.32e-07
48.5
85.95
+
8
Group 1 truncated hemoglobin
gi|17366375|sp|P73925.1|TRHBN_SYNY3
30.000
43.33
60
42
0
13
72
3
62
0.037
35.4
48.4
-
9
Methylmalonyl-CoA mutase
gi|34395931|sp|P27253.2|SCPA_ECOLI
42.105
52.63
57
26
3
21
74
19
71
2.1
31.2
7.98
-
Figure 1. Multiple alignment
Download link


Aligned length - displays length of alignment;
Identity - displays the required number of identities at a position for it to give a consensus;
Similarity - displays a cut-off for the % of positive scoring matches below which there is no consensus.
Coverage - (Aligned length / Sequence Length)* 100%
Used programs: BLAST
I've decided to use 2 randomly found proteins in the "Browse" section. After a long unsuccessful search through the families, I was able to find an interesting one.
ID Length Protein name Organism P4HA1_MOUSE (Q60715) 534 aa Prolyl 4-hydroxylase subunit alpha-1 Mus musculus (Mouse) A0A0V0VLS6_9BILA (A0A0V0VLS6) 969 aa Prolyl 4-hydroxylase subunit alpha-1 Trichinella sp. T9
Table 4. Chosen proteins
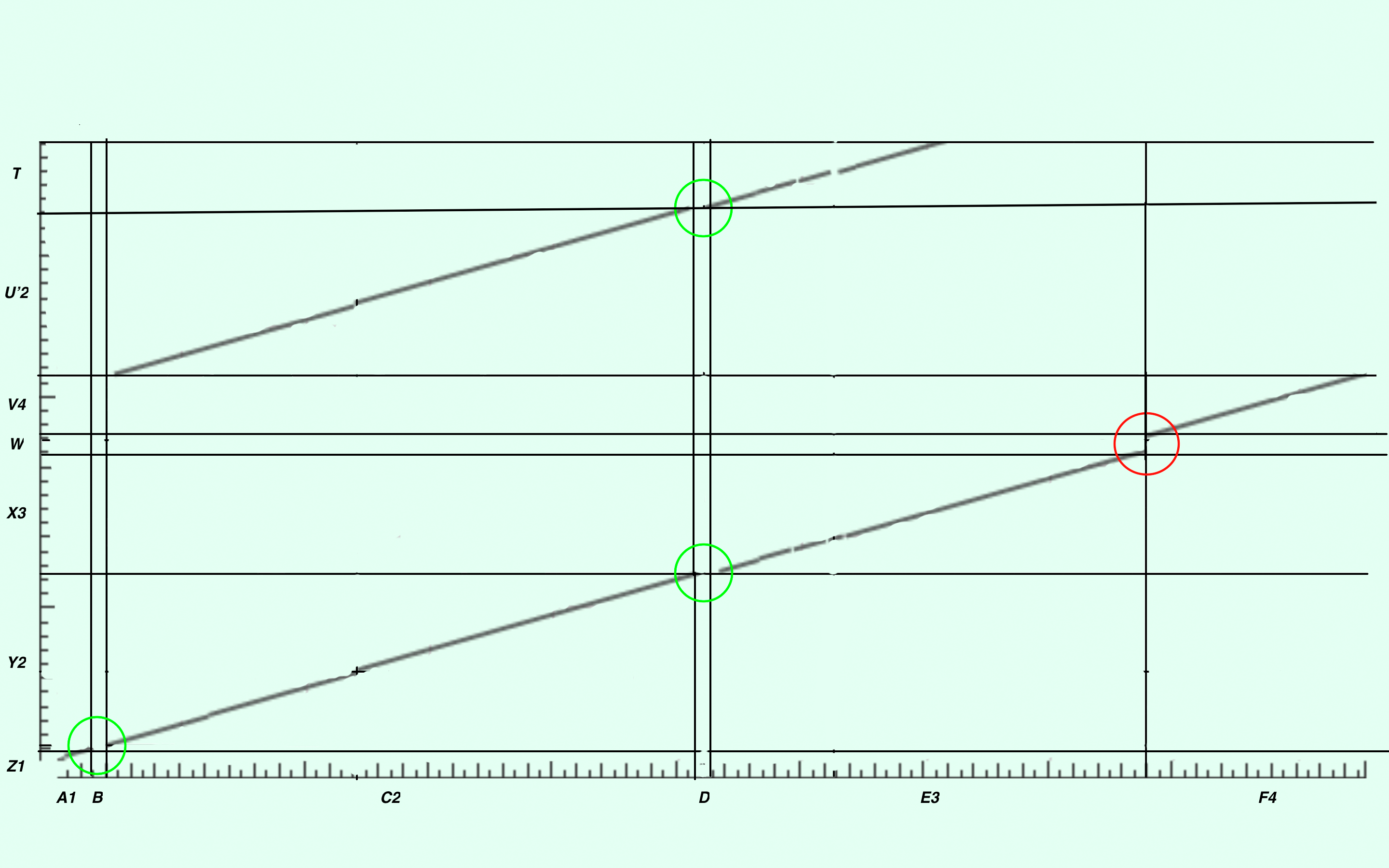
Figure 2. Map of alignment.