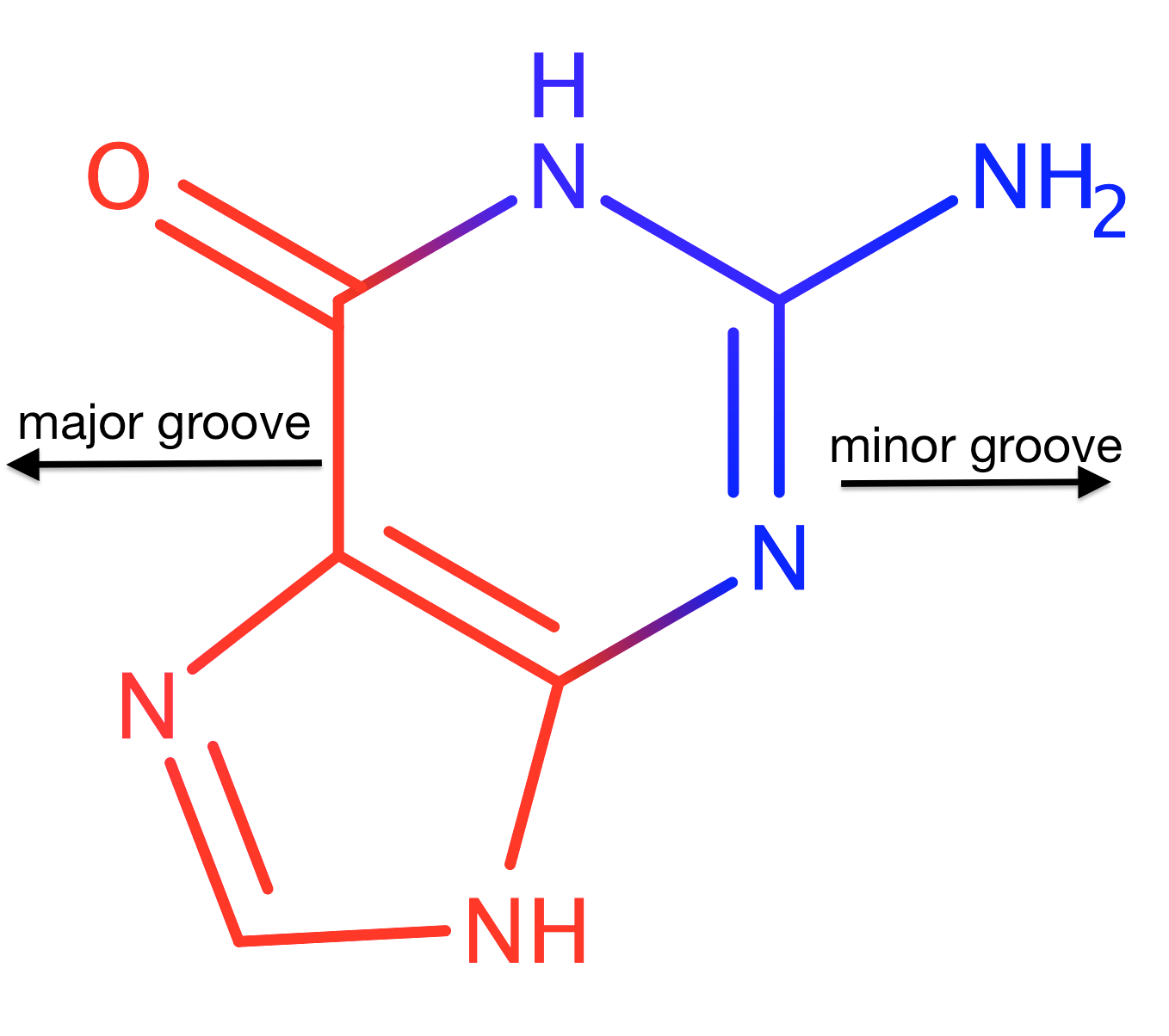
Figure 1.Marvin Sketch guanine.
Major groove | Minor groove | |
|---|---|---|
C |
C5, C6, C8 | C2, C4 |
N |
N1, N7 | N2, N3, N9 |
O |
O6 | - |
Used programms: Terminal
The task was to use fiber command in 3DNA pack. Results are in .pdb files, presented below:
Download gatc-a.pdb, gatc-b.pdb, gatc-z.pdb files ↓
|
|
|
A-form |
B-form |
Z-form |
|
|---|---|---|---|
Helical rotation (left/right) |
Right |
Right |
Left |
Pitch/helical turn (Å) |
28.03 |
33.75 |
43.5 |
Bp/turn |
11 | 10+ | 12 |
Major groove width (Å) |
16.81 | Distance [DT]31:B.P #618 - [DC]12:A.P #228 | 17.21 | Distance [DT]27:B.P #536 - [DA]10:A.P #187 | 22.91 | Distance [DC]2:A.P #23 - [DG]23:B.P #452 |
Minor groove width (Å) |
7.98 | Distance [DT]3:A.P #44 - [DC]32:B.P #638 | 11.69 | Distance [DC]12:A.P #228 - [DG]33:B.P #657 | 9.87 | Distance [DC]10:A.P #187 - [DG]23:B.P #452 |
Table 3. tRNA strand I data
|
Table 4. tRNA strand II data
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Table 5. DNA strand I data
|
Table 6. DNA strand II data
|
tRNA stems
Acceptor stem
1 (0.023) ....>C:...1_:[..G]G-----C[..C]:..72_:C<.... (0.006) |
2 (0.005) ....>C:...2_:[..C]C-----G[..G]:..71_:C<.... (0.017) |
3 (0.016) ....>C:...3_:[..C]C-----G[..G]:..70_:C<.... (0.016) |
4 (0.025) ....>C:...4_:[..G]G-----C[..C]:..69_:C<.... (0.013) |
5 (0.015) ....>C:...5_:[..A]A-----U[..U]:..68_:C<.... (0.017) |
6 (0.015) ....>C:...6_:[..G]G-----C[..C]:..67_:C<.... (0.012) |
7 (0.014) ....>C:...7_:[..G]G-----C[..C]:..66_:C<.... (0.013) |
T-stem
8 (0.006) ....>C:..49_:[..C]C-----G[..G]:..65_:C<.... (0.011) |
9 (0.016) ....>C:..50_:[..G]G-----C[..C]:..64_:C<.... (0.014) |
10 (0.015) ....>C:..51_:[..C]C-----G[..G]:..63_:C<.... (0.019) |
11 (0.011) ....>C:..52_:[..G]G-----C[..C]:..62_:C<.... (0.017) |
12 (0.015) ....>C:..53_:[..G]G-----C[..C]:..61_:C<.... (0.013) x
Anticodon stem
13 (0.014) ....>C:..39_:[..U]U-----A[..A]:..31_:C<.... (0.024) |
14 (0.010) ....>C:..40_:[..C]C-----G[..G]:..30_:C<.... (0.024) |
15 (0.020) ....>C:..41_:[..G]G-----C[..C]:..29_:C<.... (0.006) |
16 (0.017) ....>C:..42_:[..C]C-----G[..G]:..28_:C<.... (0.012) |
17 (0.014) ....>C:..43_:[..A]A-----U[..U]:..27_:C<.... (0.020) |
D-stem
19 (0.014) ....>C:..10_:[..G]G-----C[..C]:..25_:C<.... (0.015) |
20 (0.012) ....>C:..11_:[..C]C-----G[..G]:..24_:C<.... (0.025) |
21 (0.013) ....>C:..12_:[..U]U-----A[..A]:..23_:C<.... (0.015) |
22 (0.010) ....>C:..13_:[..C]C-----G[..G]:..22_:C<.... (0.019) |
Stabilising h-bonds
23 (0.019) ....>C:..14_:[..A]A-**--U[..U]:...8_:C<.... (0.006) |
24 (0.012) ....>C:..15_:[..G]G-**+-C[..C]:..48_:C<.... (0.031) |
25 (0.011) ....>C:..16_:[..U]U-**+-U[..U]:..59_:C<.... (0.016) x
26 (0.019) ....>C:..18_:[..G]G-**+-U[..U]:..55_:C<.... (0.009) |
27 (0.026) ....>C:..19_:[..G]G-----C[..C]:..56_:C<.... (0.014) x
28 (0.013) ....>C:..73_:[..A]A-**+-C[..C]:..74_:C<.... (0.017) +
29 (0.016) ....>C:..45_:[..U]U-**--A[..A]:...9_:C<.... (0.017) +
7 non-Watson-Crick base-pairs
18 (0.019) ....>C:..44_:[..G]G-**--A[..A]:..26_:C<.... (0.022) |
23 (0.019) ....>C:..14_:[..A]A-**--U[..U]:...8_:C<.... (0.006) |
24 (0.012) ....>C:..15_:[..G]G-**+-C[..C]:..48_:C<.... (0.031) |
25 (0.011) ....>C:..16_:[..U]U-**+-U[..U]:..59_:C<.... (0.016) x
26 (0.019) ....>C:..18_:[..G]G-**+-U[..U]:..55_:C<.... (0.009) |
28 (0.013) ....>C:..73_:[..A]A-**+-C[..C]:..74_:C<.... (0.017) +
29 (0.016) ....>C:..45_:[..U]U-**--A[..A]:...9_:C<.... (0.017) +
|
The 23, 24, 29 bp are also non-Watson-Crick despite their "common" pairing. that is because of a different kind of interaction between them. In the Fig.2 there is an exapmle of such pair:
|