| Organism: Helicobacter pylori |  |
| Data received from NPG explorer (kodomo) | |||||
|---|---|---|---|---|---|
| all blocks | s-blocks | h-blocks | r-blocks | m-blocks | |
| Amount | 619 | 261 | 220 | 138 | 440 |
| Min identity | 0.7866 | 0.7866 | 0.8138 | 0.8305 | 0 |
| Max identity | 1 | 0.9787 | 1 | 1 | 1 |
| Avg identity | 0.9301 | 0.9349 | 0.9342 | 0.9145 | 0.6867 |
| Med identity | 0.9397 | 0.9492 | 0.9391 | 0.9044 | 0.8355 |
| Coverage, % | 89.24 | 79.41% | 7.3% | 2.52% | 0.75% |
| Received images : | |
|---|---|
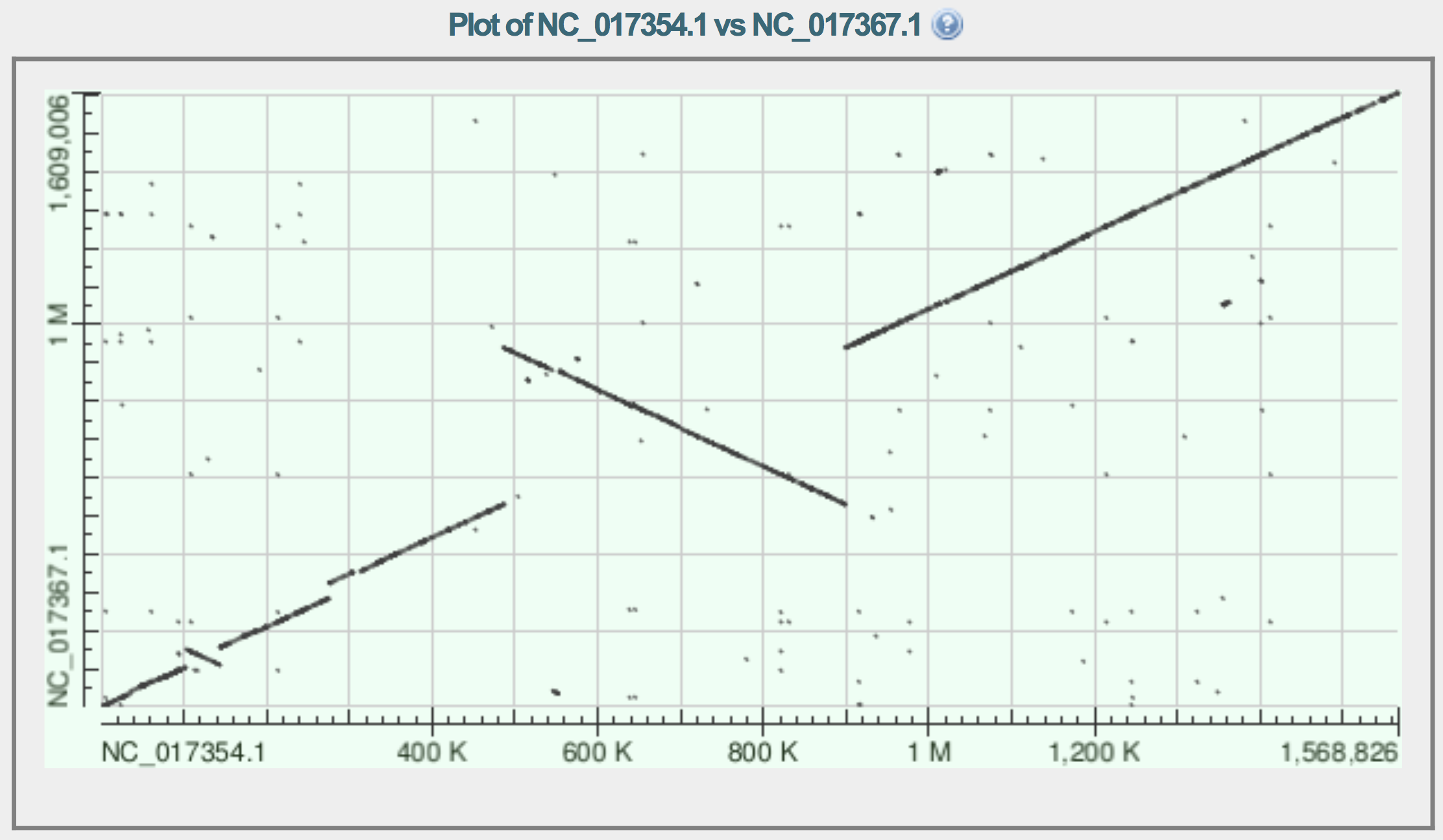 | Inversion example (480K-900K, axis x) Inversion example (100K-140K, axis x) |
 | Inversion example (480K-900K, axis x) Inversion + translocation example (20K-60K, axis x) |
 | Inversion + translocation example (20K-60K, axis x) Translocation example (280K-320K, axis x) |