Sanger sequencing
Sanger sequencing
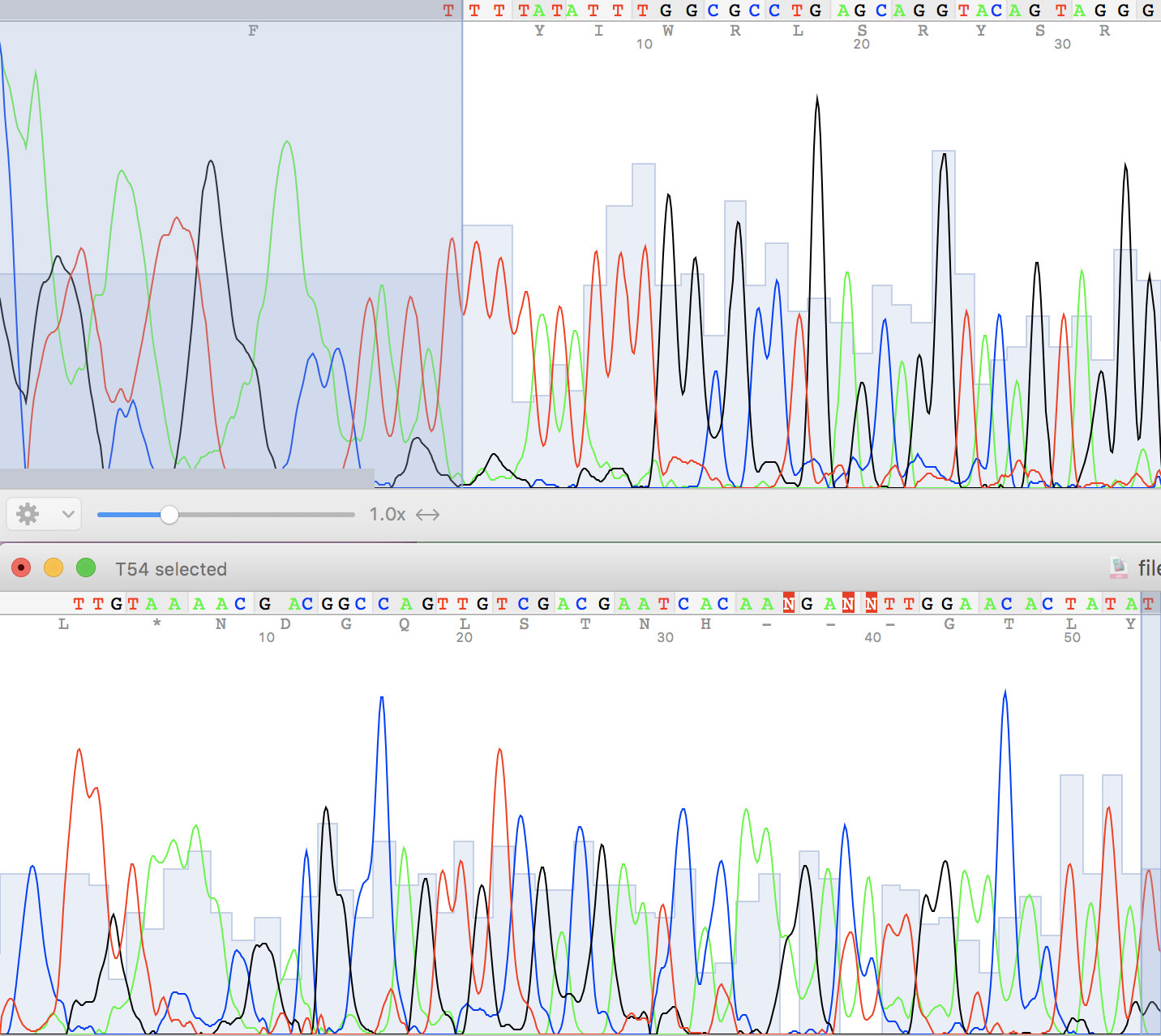
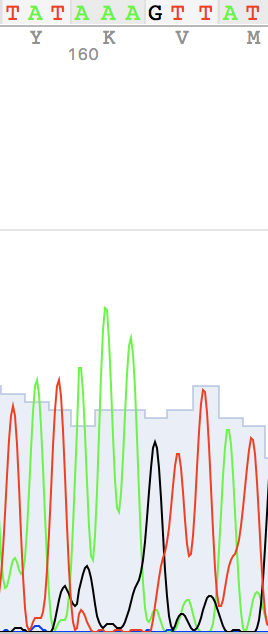
Brief chromatogram description
Non-readable regions:
Direct sequence
Reverse sequence
5'
1-23
1-2
3'
705-719
687-718
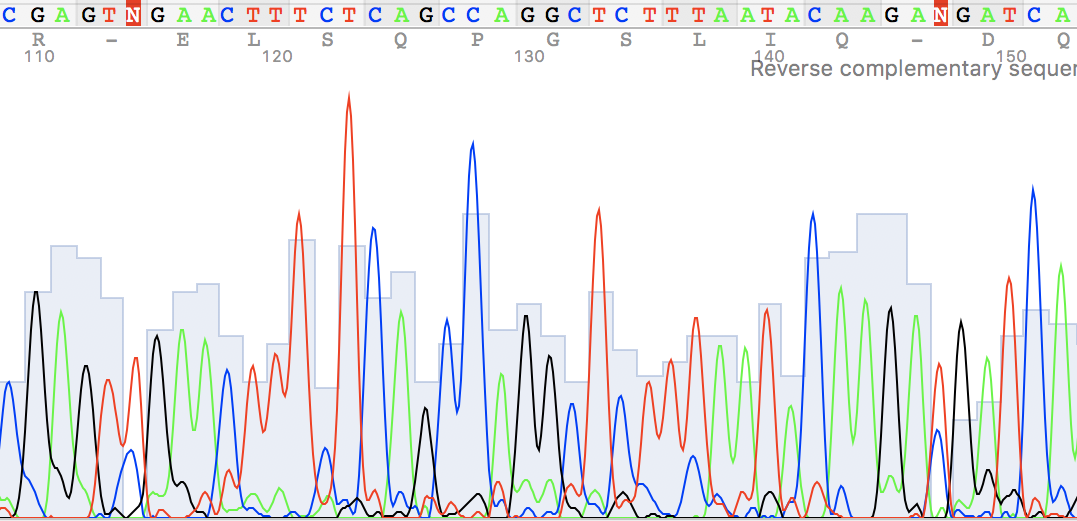
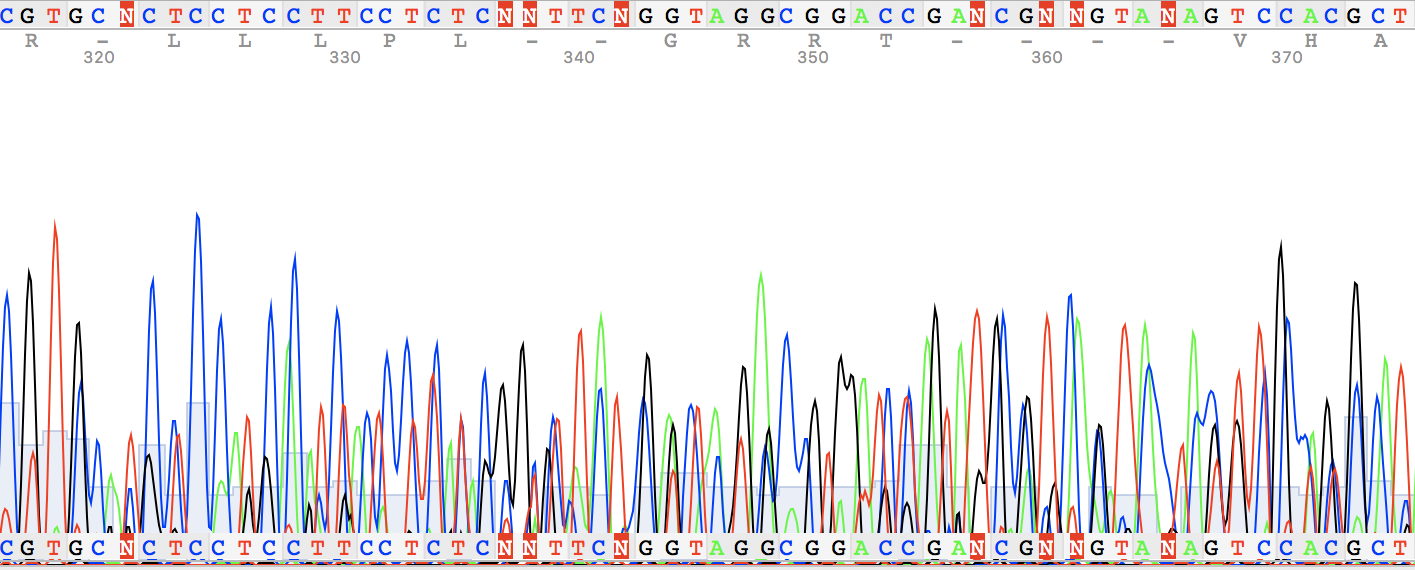
Figure 1.
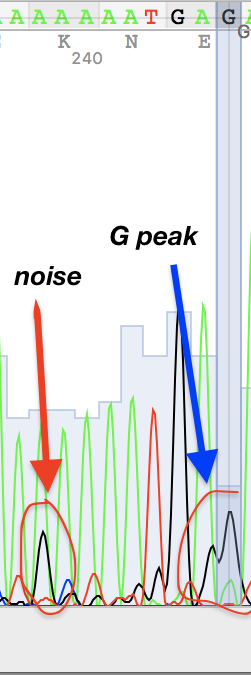
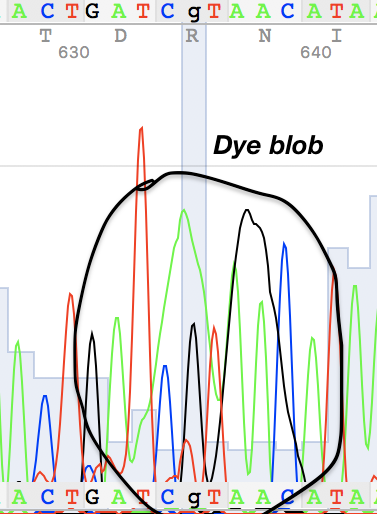
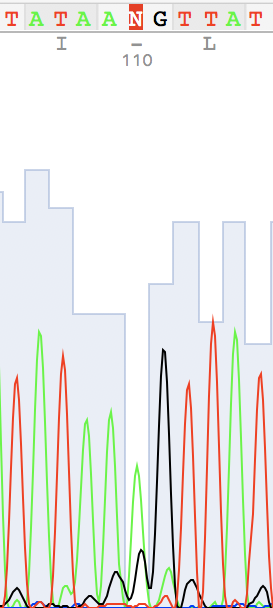
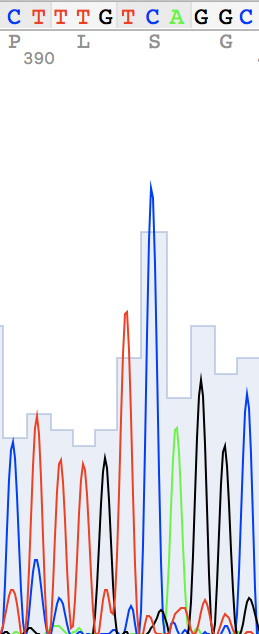
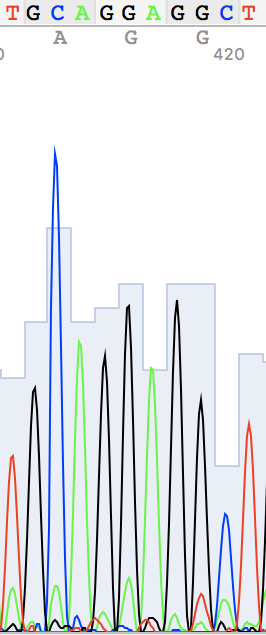
I've also found two unusual regions:
Figure 2. Figure 3.
Jalview alignment
{"seqs":[{"name":"file1/1-681","start":1,"end":681,"id":"301528529","seq":"-----------------------------------------------------TTTTATATTTGGCGCCTGAGCAGGTACAGTAGGGACTGCCATGAGAAAAATTATACGAGTTGAACTTTCTCAGCCAGGCTCTTTAATACAAGATGATCAAGTATATAAaGTTATGGTAACGGCCCACGCCTTCGTCATGATATTTTTTATGGTAATGCCCATAATGATAGGGGGGTTTGGCAAATGACTTGTCCCACTAATGTTAGGAGCGCCkGATATGGCTTTCCCCCGAATGAAAAAAATGAGATTTTGGCTACTACCCCCAGCTTTTATACTTCTTCTAGCTTCAGCTGCAAACGAAGGAGGAGTAGGCACTGGATGAACTATTTATCCCCCTTTGtCAGGCCCTACCGCACATGCAGgAGGCTGCGTAGACCTCGCAATTTTTTCTCTCCACCTAGCAGGTGCGTCTTCAATTATGGCCTCAATAAAATTTATTACAACTATwATAAATATGCGTAGGCCCGGCATGACCATGGATCGACTTCCACTTTTTGCTTGATCTATTTTCTTAACAACTATATTACTACTCCTTTCTCTGCCTGTTTTAGCAGGAGCTATTACAATGCTATTAACTGATCGTAACATAAAAACAACGTTTTTTGATCCTACAGGAGGAGGAGACCCAATACTTTTCCAACATTtATTTTGrTTyTTyGGCCAyCCCGAGGtCTAGTCATA","order":1},{"name":"file2/1-684","start":1,"end":684,"id":"1252393673","seq":"TTGTAAAACGACGGCCAGTTGTCGACGAATCACAArGAhhTTGGAACACTATATTTTATATTTGGCGCCTGAGCAGGTACAGTAGGGACTGCCATGAGAAAAATTATACGAGTyGAACTTTCTCAGCCAGGCTCTTTAATACAAGAyGATCAAGTATATAAAGTTATGGTAACGGCCCACGCCTTyGTCaTGATATTTTTTATGGTAATGCCCATAATGATAGGGGGGTTTGGCAAATGACTTGTCCCACTAATGTTAGGAGCGCCTGATATGGCTTTCCCCCGAATGAAAAAAATGAGATTTTGGCTACTACCCCCAGCTTtTATACTTCTTCTAGCTTCAGCTGCAAACGAAGGAGGAGTAGGCACTGGATGAmCTATTTATCCCCCTTTGTCAGGCCCTACCGCACATGCAGGAGGCTGCGTAGACCTCGCAATTTTTTCTCTCCmCCTAGCAGGTGCGTCTTCAATTATGGCCTCAATAAAATTTAyTACAACTATTATAAATATGCGTAGGCCCGGCATGACCATGGATCGACTTCCACTTTTTGCTTGATCTATTTTCTTAACAACTATATTACTACTCCTTTCTCTGCCTGTTTTAGCAGGAGCTATTACAATGCTATTAACTGATCgTAACATAAAAACAACGTTTTTTGATCCTACAGGAGGAGGaGACCCAATA--------------------------------------------------","order":2}],"appSettings":{"globalColorScheme":"Clustal","webStartUrl":"http://www.jalview.org/services/launchApp","application":"Jalview","showSeqFeatures":"false","version":"2.10.1"},"seqGroups":[],"alignAnnotation":[],"svid":"1.0","seqFeatures":[]}
10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 180 190 200 210 220 230 240 250 260 270 280 290 300 310 320 330 340 350 360 370 380 390 400 410 420 430 440 450 460 470 480 490 500 510 520 530 540 550 560 570 580 590 600 610 620 630 640 650 660 670 680 690 700 710 720 730 - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - T T T T A T A T T T G G C G C C T G A G C A G G T A C A G T A G G G A C T G C C A T G A G A A A A A T T A T A C G A G T T G A A C T T T C T C A G C C A G G C T C T T T A A T A C A A G A T G A T C A A G T A T A T A A a G T T A T G G T A A C G G C C C A C G C C T T C G T C A T G A T A T T T T T T A T G G T A A T G C C C A T A A T G A T A G G G G G G T T T G G C A A A T G A C T T G T C C C A C T A A T G T T A G G A G C G C C k G A T A T G G C T T T C C C C C G A A T G A A A A A A A T G A G A T T T T G G C T A C T A C C C C C A G C T T T T A T A C T T C T T C T A G C T T C A G C T G C A A A C G A A G G A G G A G T A G G C A C T G G A T G A A C T A T T T A T C C C C C T T T G t C A G G C C C T A C C G C A C A T G C A G g A G G C T G C G T A G A C C T C G C A A T T T T T T C T C T C C A C C T A G C A G G T G C G T C T T C A A T T A T G G C C T C A A T A A A A T T T A T T A C A A C T A T w A T A A A T A T G C G T A G G C C C G G C A T G A C C A T G G A T C G A C T T C C A C T T T T T G C T T G A T C T A T T T T C T T A A C A A C T A T A T T A C T A C T C C T T T C T C T G C C T G T T T T A G C A G G A G C T A T T A C A A T G C T A T T A A C T G A T C G T A A C A T A A A A A C A A C G T T T T T T G A T C C T A C A G G A G G A G G A G A C C C A A T A C T T T T C C A A C A T T t A T T T T G r T T y T T y G G C C A y C C C G A G G t C T A G T C A T A T T G T A A A A C G A C G G C C A G T T G T C G A C G A A T C A C A A r G A h h T T G G A A C A C T A T A T T T T A T A T T T G G C G C C T G A G C A G G T A C A G T A G G G A C T G C C A T G A G A A A A A T T A T A C G A G T y G A A C T T T C T C A G C C A G G C T C T T T A A T A C A A G A y G A T C A A G T A T A T A A A G T T A T G G T A A C G G C C C A C G C C T T y G T C a T G A T A T T T T T T A T G G T A A T G C C C A T A A T G A T A G G G G G G T T T G G C A A A T G A C T T G T C C C A C T A A T G T T A G G A G C G C C T G A T A T G G C T T T C C C C C G A A T G A A A A A A A T G A G A T T T T G G C T A C T A C C C C C A G C T T t T A T A C T T C T T C T A G C T T C A G C T G C A A A C G A A G G A G G A G T A G G C A C T G G A T G A m C T A T T T A T C C C C C T T T G T C A G G C C C T A C C G C A C A T G C A G G A G G C T G C G T A G A C C T C G C A A T T T T T T C T C T C C m C C T A G C A G G T G C G T C T T C A A T T A T G G C C T C A A T A A A A T T T A y T A C A A C T A T T A T A A A T A T G C G T A G G C C C G G C A T G A C C A T G G A T C G A C T T C C A C T T T T T G C T T G A T C T A T T T T C T T A A C A A C T A T A T T A C T A C T C C T T T C T C T G C C T G T T T T A G C A G G A G C T A T T A C A A T G C T A T T A A C T G A T C g T A A C A T A A A A A C A A C G T T T T T T G A T C C T A C A G G A G G A G G a G A C C C A A T A - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
Download files:
Original data: direct ↓ and reverse ↓
FASTA: ↓
JalView alignment: ↓
My edits
#
1
2
3
4 and 5
Direct sequence
Figure 4a. Figure 5a. Figure 6a. Figure 7a.
Reverse sequence
Figure 4b. Figure 5b. Figure 6b. Figure 7b.
Description
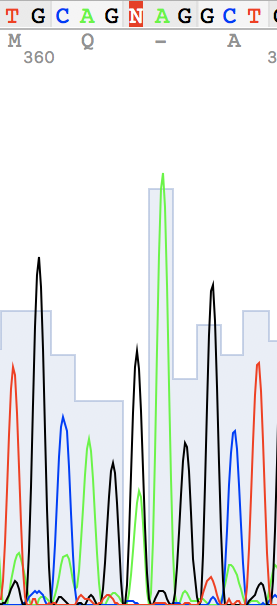
Unidentified nucleotide (N #341) was replaced to T (according to thymine #394 in the reverse sequence) N #364 in direct replaced to G (see reverse guanine #416) N #110 in direct replaced to A (see reverse adenine #162) N #114 in reverse replaced to T (see direct thymine #62)
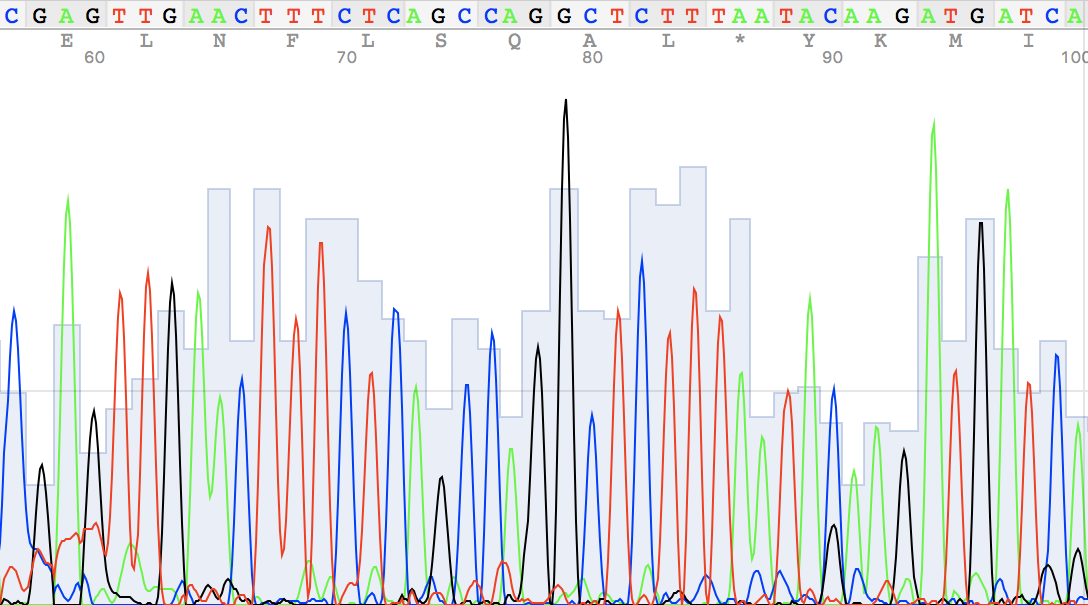
An example of a non-readable chromatogram
Figure 8. Back to term 3 page 🚶