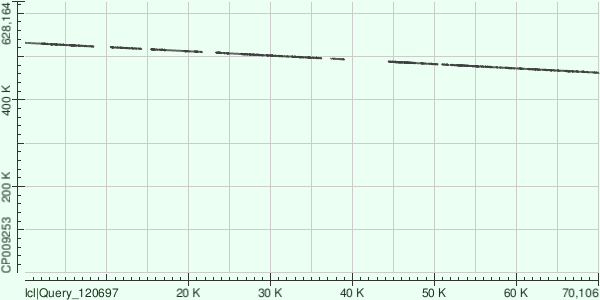

| Contig | Genome |
|---|---|
| [57822:65135] | [474667:467412] |
| [5:2796] | [531590:528794] |
| [23323:31768] | [508806:500370] |
| [15452:21628] | [516539:510438] |
| [2866:8442] | [528679:523105] |
| [65162:70106] | [467421:462496] |
| [44408:50485] | [488106:481997] |
| [51845:57719] | [480660:474844] |
| [10457:14186] | [521500:517766] |
| [31884:36161] | [500325:496111] |
| [37579:38955] | [494864:493487] |
| [50957:51639] | [481545:480874] |
| [37327:37445] | [495148:495033] |
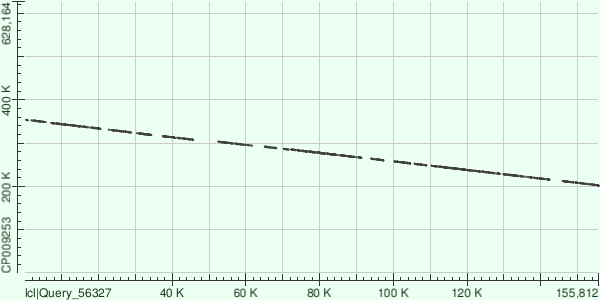
| Contig | Genome |
|---|---|
| [81937:91416] | [275551:266073] |
| [110407:121104] | [247596:236918] |
| [12176:20526] | [341508:333222] |
| [52503:59856] | [303252:295935] |
| [73657:81878] | [283706:275566] |
| [150564:155812] | [207661:202390] |
| [134400:138502] | [223720:219625] |
| [41358:45669] | [312179:307878] |
| [129900:134011] | [228137:224057] |
| [121202:125731] | [236859:232358] |
| [125880:129000] | [232057:228944] |
| [93966:97537] | [263784:260224] |
| [6511368479] | [291560:288181] |
| [105728:108930] | [252161:248967] |
| [37405:40713] | [315982:312679] |
| [139649:142334] | [218384:215717] |
| [7060:10399] | [346547:343228] |
| [30158:34371] | [323043:318826] |
| [145910:148869] | [212243:209294] |
| [100315:104637] | [257546:253223] |
| [23571:26340] | [330003:327227] |
| [60165:61691] | [295755:294227] |
| [1672:4246] | [352456:349918] |
| [26632:28838] | [326950:324747] |
| [4495:5937] | [349674:348233] |
| [37:843] | [353822:353014] |
| [10545:11817] | [343052:341781] |
| [70269:71603] | [286535:285200] |
| [22517:23190] | [331006:330333] |
| [72300:73409] | [285070:283963] |
| [149315:150205] | [208904:208017] |
| [138550:139210] | [219491:218821] |
| Contig | Genome |
|---|---|
| [62724 :72165] | [398726:389348] |
| [57496 :62479] | [403823:398904] |
| [43342 :47936] | [417677:413081] |
| [14422 :17449] | [445895:442877] |
| [7198 :11850] | [454069:449411] |
| [31369 :35447] | [429483:425412] |
| [55085 :57253] | [406218:404050] |
| [22353 :25276] | [438139:435267] |
| [25338 :28767] | [435241:431839] |
| [49023 :50837] | [412321:410512] |
| [17579 :19254] | [442817:441135] |
| [74888 :76355] | [386887:385425] |
| [39557 :40410] | [421327:420477] |
| [76419 :77702] | [385420:384182] |
| [19376 :19557] | [440944:440755] |
| [19647 :19728] | [440732:440652] |
Evalue range: from 9,88E-163 to 0.0
Strand: +/-
Evalue range: from 9,88E-163 to 0.0
Strand: +/-
Evalue range: from 1.92e-165 to 0.0
Strand: +/-
Assuming data above, the whole lentgth of contig is aligned (reversed). There are also some gaps.
Assuming data above, the whole length of contig is aligned (reversed). There are also some gaps.
Assuming data above, almost whole length of contig (except 1-7K region, see hit table) is aligned (reversed). There are also some gaps.