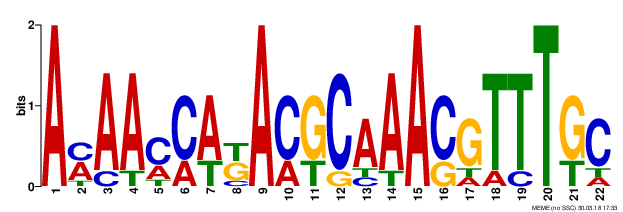
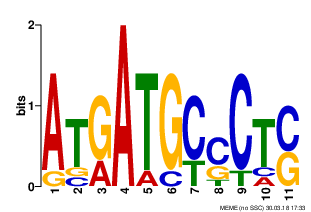
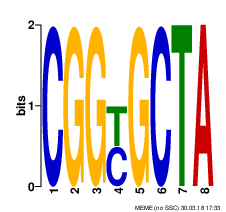| Q87Q53 | PURT_VIBPA | Formate-dependent phosphoribosylglycinamide formyltransferase | purT | complement(1376818..1377993) | complement(1377994-1378094) |
| P40607 | PURA_VIBPA | Adenylosuccinate synthetase | purA | complement(2975587..2976903) | complement(2976904 - 2977004) |
| Q87RB8 | FOLD_VIBPA | Bifunctional protein FolD | folD | complement(911485..912345) | complement(912346 - 912446) |
| Q87KE0 | PURK_VIBPA | N5-carboxyaminoimidazole ribonucleotide synthase | purK | 3244219..3245352 | 3244118 - 3244218 |
| Q87QW9 | PURR_VIBPA | HTH-type transcriptional repressor PurR | purR | complement(1079009..1080013) | complement(1080014 - 1080114) |
| Q87MH0 | PUR5_VIBPA | Phosphoribosylformylglycinamidine cyclo-ligase | purM | 2393407..2394447 | 2393306 - 2393406 |
| Q87KE1 | PURE_VIBPA | N5-carboxyaminoimidazole ribonucleotide mutase | purE | 3243729..3244214 | 3243628 - 3243728 |
| Q87Q87 | PUR7_VIBPA | Phosphoribosylaminoimidazole-succinocarboxamide synthase | purC | complement(1337584..1338687) | complement(1338688 - 1338788) |