| # | Amount of sequences producing significant alignments with E-value higher than 0,005 | ID of a sequence producing the best alignment with E-value WORSE than threshold | E-value | ID of a sequence producing the worst alignment with E-value BETTER than threshold | E-value |
| 1 | 31 | P26983.1 | 0.001 | P28613.2 | 6e-04 |
| 2 | 46 | P9WMA8.1 | 0.009 | P71346.3 | 5e-11 |
| 3 | 51 | P9WMA8.1 | 0.002 | P24694.1 | 2e-23 |
| 4 | 51 | P9WMA8.1 | 0.002 | P24694.1 | 5e-24 |
| 5 | 53 | P9WMA8.1 | 0.001 | P24694.1 | 4e-24 |
| 6 | 53 | P9WMA8.1 | 0.001 | P24694.1 | 5e-24 | Next iterations had the same amount of sequences. |
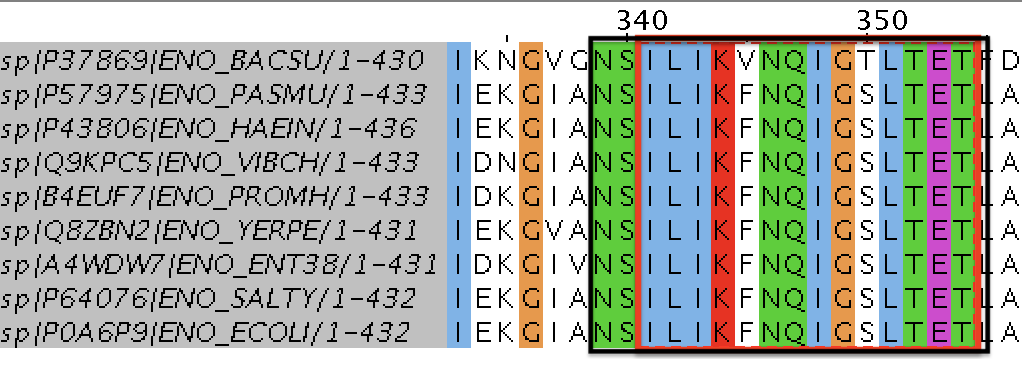
Then, using PROSITE, I scanned my changed pattern and found 296 hits. In Uniprot list there are, however, 396 hits.
File with my hits: pattern.txt
Uniprot file: true.txt
To find all differences between those two lists, the Python script was created and all results were written down in the table below:
| TP (True Positives - presented in both lists) | 148 |
| FP (False Positives - not presented in Uniprot list) | 148 |
| FN (False Negatives - hits from Uniprot list which weren't found in my list from PROSITE) | 248 |