Domains. Pfam. Profile HMM.
Domains. Pfam. Profile HMM.
Task 1
AC: PF01237
ID: Oxysterol_BP
Domain function: lipid-binding, implicated in many cellular processes related with oxysterol, including signaling, vesicular trafficking, lipid metabolism, and nonvesicular sterol transport.
Pfam link
Domain architecture link
Chosen domain architectures:
| Image |
3D structure |
Amount of sequences |
Description |
 |
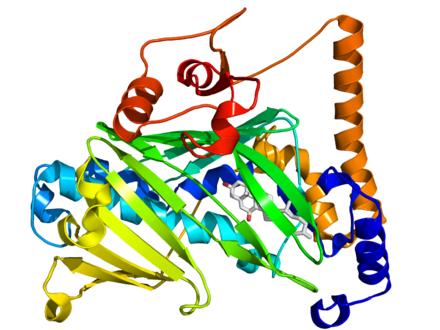 |
2639 |
Contains only one domain. |
 |
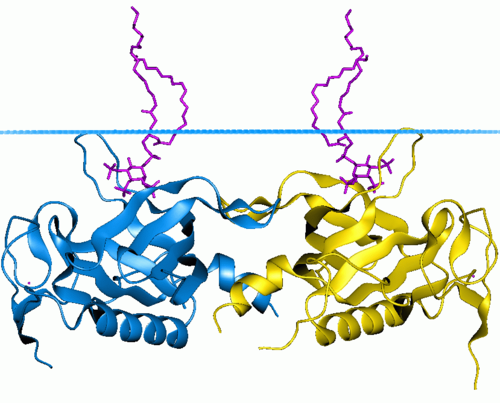 |
677 |
Pleckstrin homology domain function:
This domain can bind phosphatidylinositol lipids within biological membranes and proteins. Through different interactions,
PH domain plays an important role in
recruiting proteins to different membranes, thus targeting them to appropriate cellular compartments or enabling them to interact
with other components of the signal
transduction pathways.
|
Chosen taxon: Eukaryota
Chosen subtaxa: Fungi, Metazoa
Download Excel table
Using the third .py script and Jalview,
I was able to create .jvp file with 2 groups (as for 2 domain architectures) with 40 seqeunces (20 from Fungi, 20 from Metazoa) in each of them.
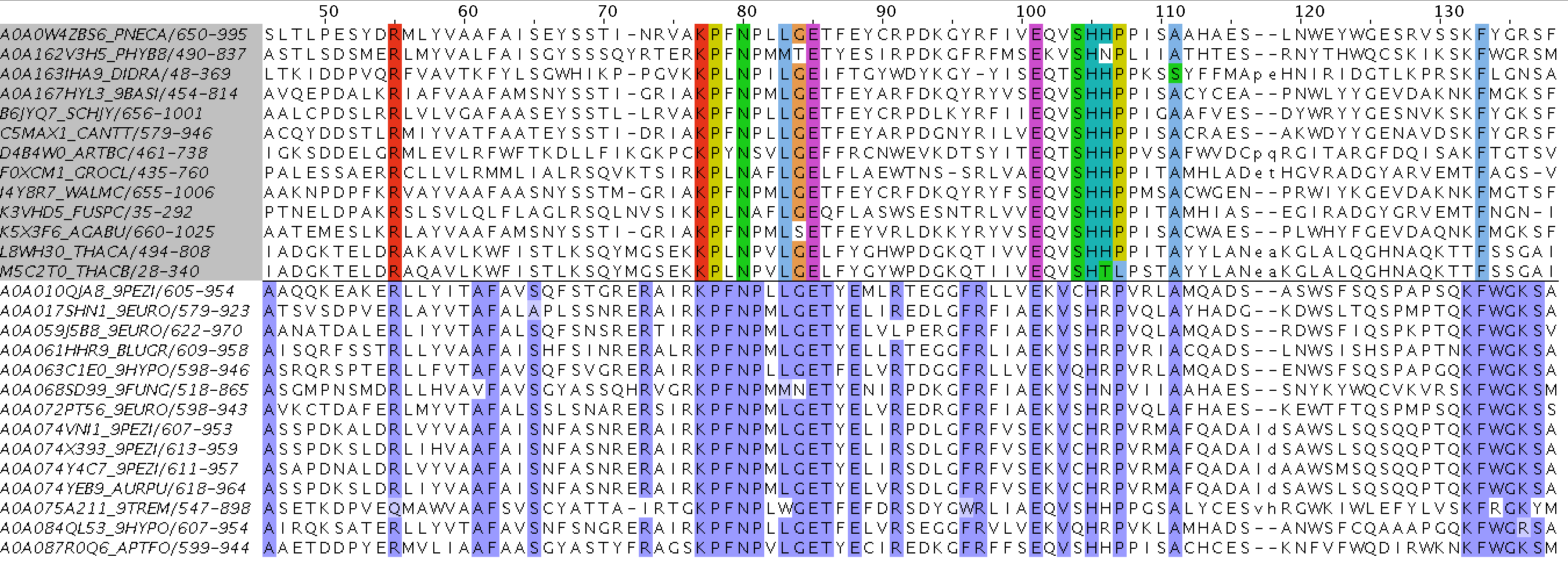
A part of my alignment
Download Jalview alignment
Even from the picture above could be noticed that the second group sequences are more conservative (conservation threshold was set to 100% for both groups).
What is more, many conservative columns from the first group and the second are the same because of a similar structure.
Task 2
Using MEGA, I constructed maximum likelihood tree. 1, 2 stand for domain architectures, F for Fungi, M for Metazoa.
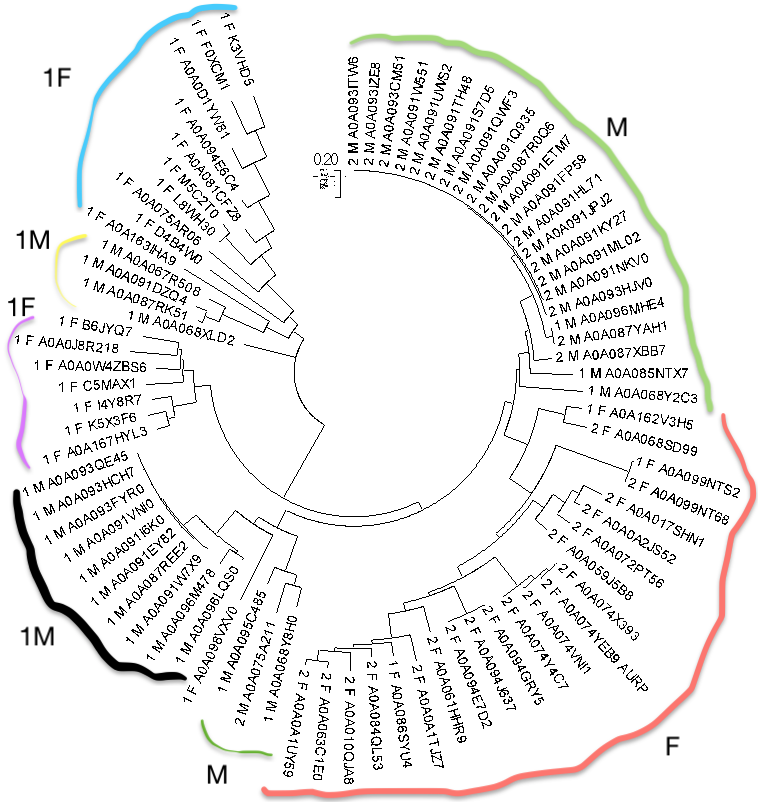 |
WIth coloured lines I marked similar groups, sometimes clades (e.g light green M, which is next to light blue F). A division by domain architecures is not strict, but quite discernible: left part is almost all 1, while the right contains mostly 2's, it means that the architectures were formed before taxons because there is no strict division (left-right i mean) in Metazoa-Fungi taxons.
Download tree formula |
Back to term 4 page 🚶
© Sophia Veselova, 2018.



