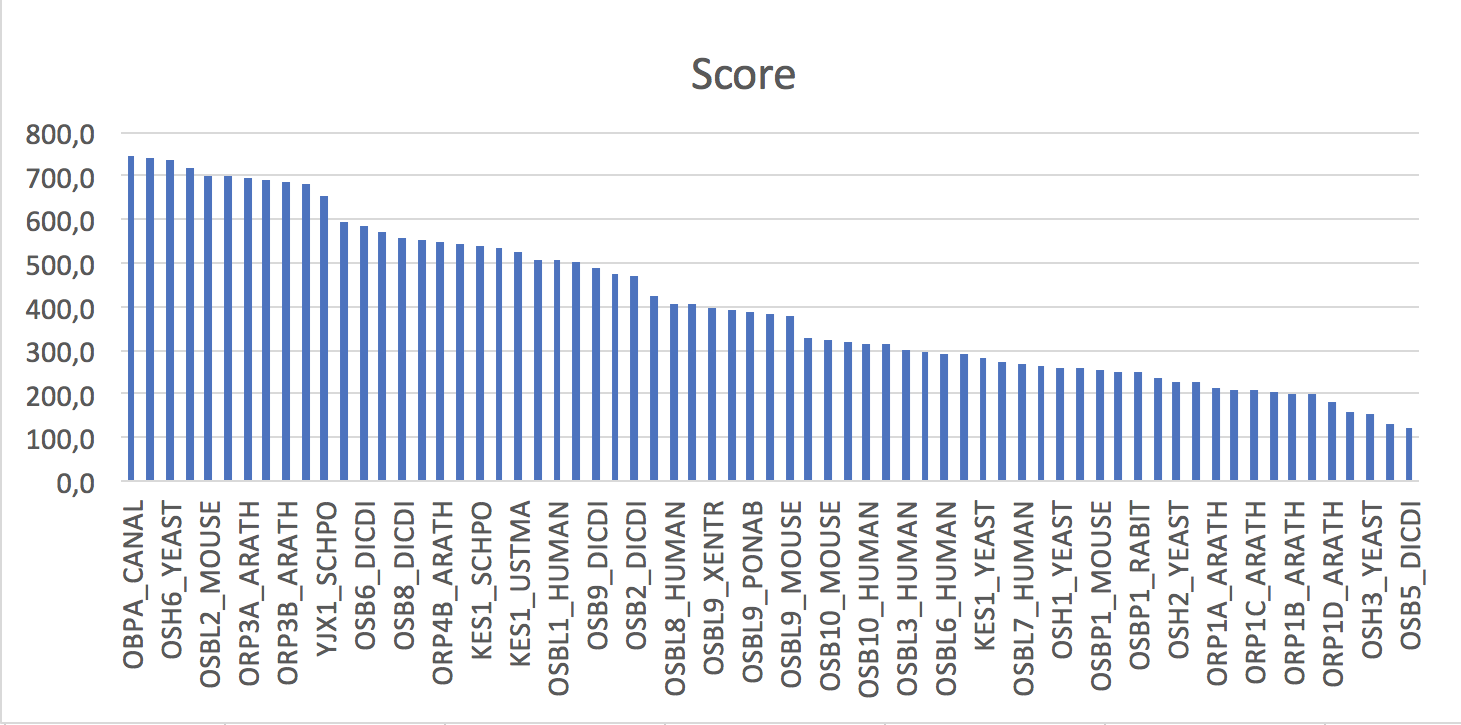
Fig. 2. Created histogram
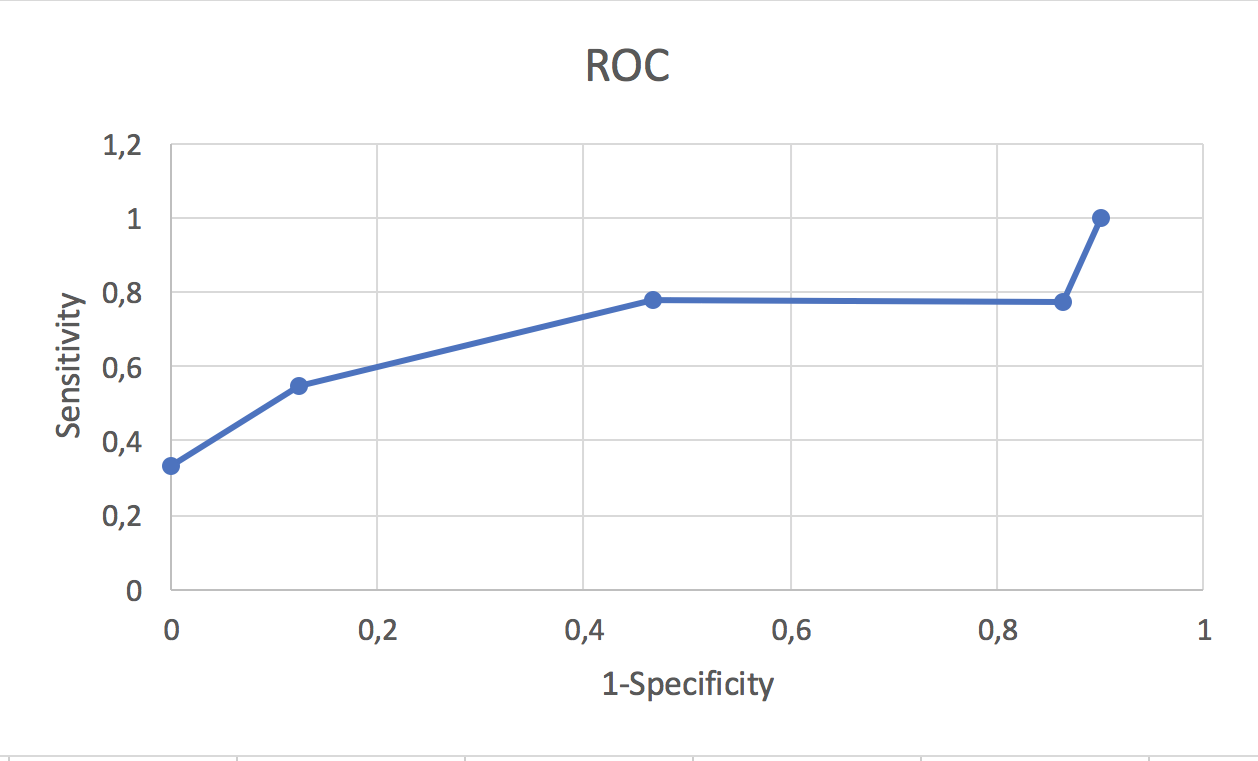
Fig. 3. ROC-curve

Fig. 2. Created histogram |

Fig. 3. ROC-curve |
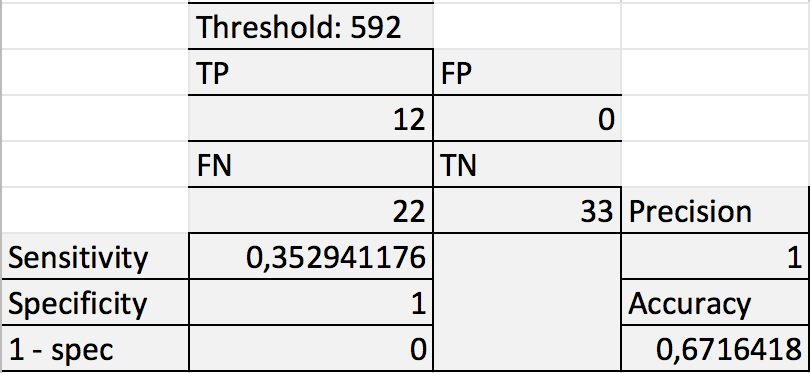
Fig. 4. Data for chosen threshold |
As we can see, ACU - Area Under Curve - isn't equal to 1, which means my prediction isn't perfect, but okay.
Precision and Recall are 1 (and that's great - everything i found is in), but the Accuracy value is only 67%. |