|
Home page Term 1 Term 2 Term 3 About me Faculty website |
Genome BrowsersUCSCThe protein properdin was chosen for this task.
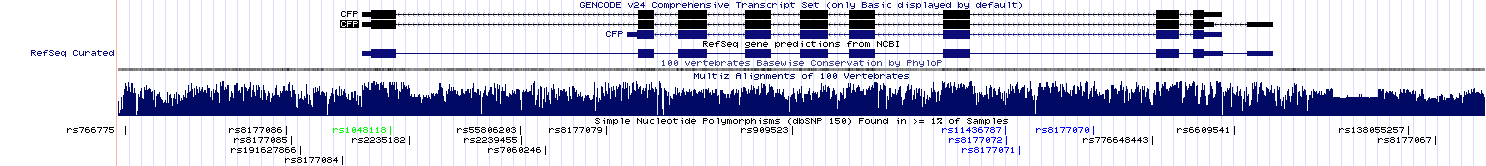
ENSEMBLThe distmat program gave the distance of 0.67, meaning that, on average, there are 0.67 substitutions per every 100 bases. ENSEMBL shows the length of the aligned human sequence to be 8333 bp; the number of substitutions is, consequently, 0.67 * (8333 / 100) = 55.8 (more likely to be 56).Alignment After setting the location filter close to the coordinates of the aligned sequence, the variant table showed the overall number of SNPs to be 31539. 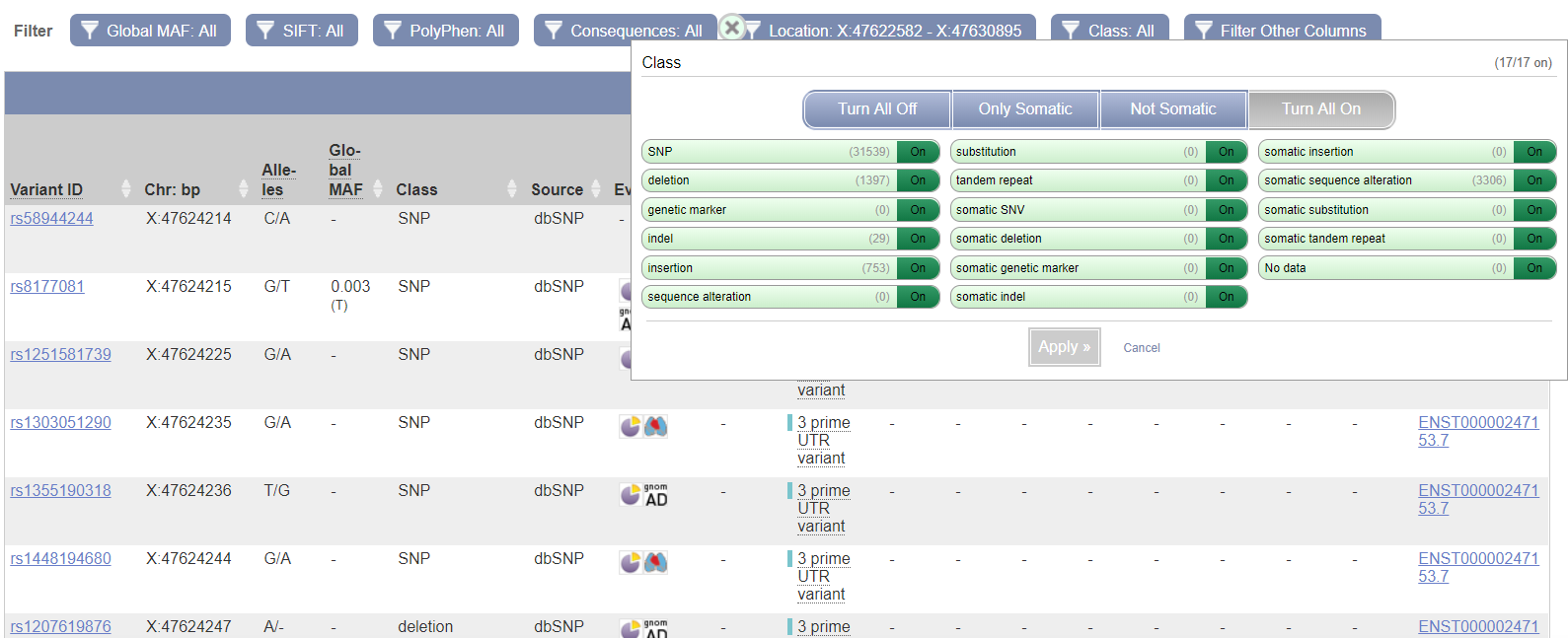 To conclude, humans are indeed diverse, but such a great difference stems from the fact that only one human sequence was aligned with only one chimpanzee sequence, though chimpanzees also have sequence variants. NCBI Genome Data ViewerOne advantage of this browser is that each exon can be highlighted in all the displayed tracks simultaneously (exon navigation). Also, there is a picture with all the human chromosomes, which can be used for navigation.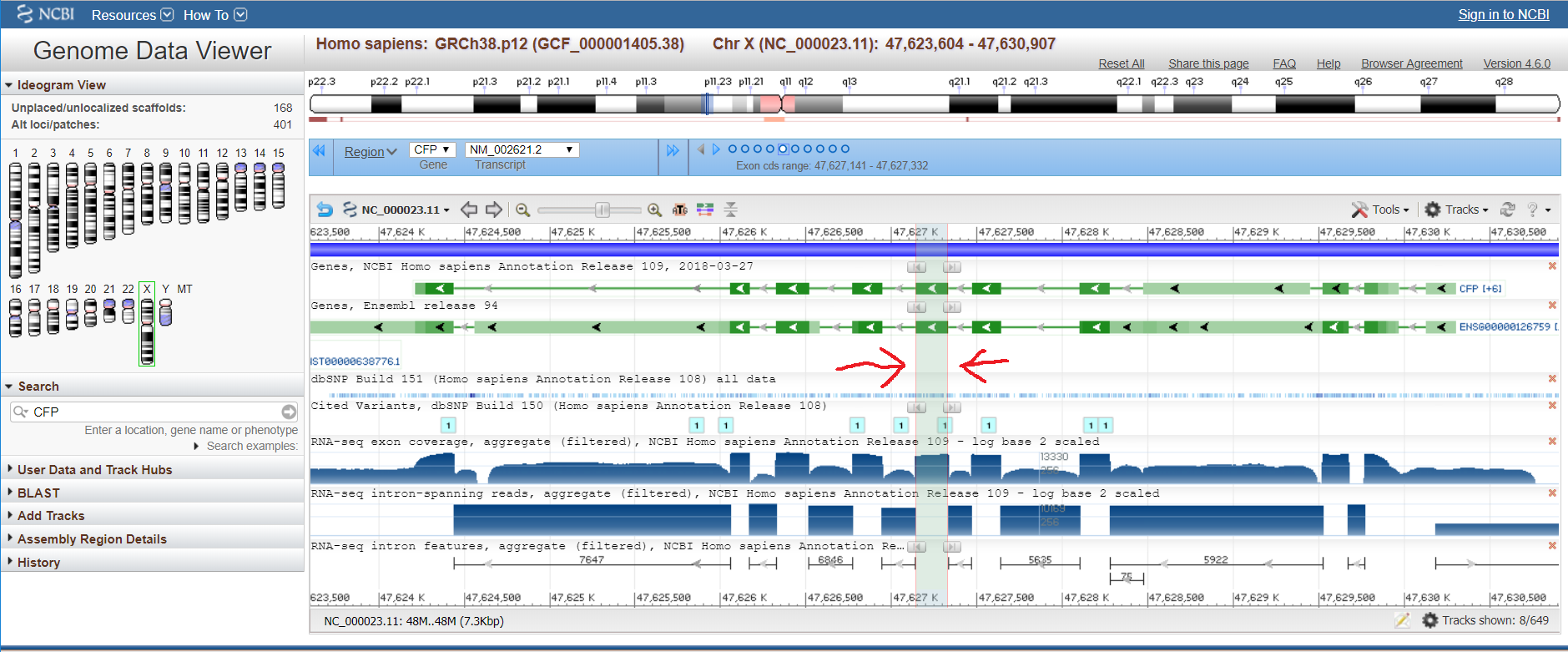
|
© Stanislav Tikhonov, 2018